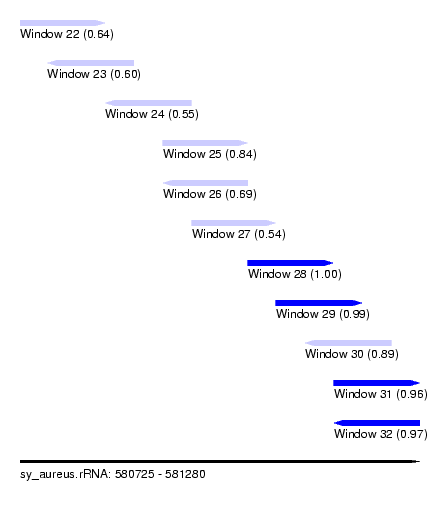
| Sequence ID | sy_aureus.rRNA |
|---|---|
| Location | 580,725 – 581,280 |
| Length | 555 |
| Max. P | 0.999568 |

| Location | 580,725 – 580,843 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.87 |
| Mean single sequence MFE | -36.78 |
| Consensus MFE | -35.75 |
| Energy contribution | -32.62 |
| Covariance contribution | -3.12 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.637198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 580725 118 + 2809430 GCUAGCCUCAA-GUG-AUGAUUAUUGGAGGUAGAGCACUGUUUGGACGAGGGGCCCCUCUCGGGUUACCGAAUUCAGACAAACUCCGAAUGCCAAUUAAUUUAACUUGGGAGUCAGAACA .((..((((((-((.-..(((((((((.....(((...((((((((.((((....))))((((....)))).))))))))..)))......)))).)))))..))))))).)..)).... ( -39.30) >sp_agal.rRNA 19102 119 + 2160275 GCUAGCGUCGAUGUU-AAGUCUCUUGGAGGUAGAGCACUGUUUGGGUGAGGGGUCCAUCCCGGAUUACCAAUCUCAGAUAAACUCCGAAUGCCAACGAGAUAUAAUCGGCAGUCAGACUG .((..(((((((...-..(((((((((.....(((...((((((((...(((((((.....))))).))...))))))))..)))......)))).)))))...)))))).)..)).... ( -34.90) >sp_mutans.rRNA 19682 119 + 2030929 GCUAGCGUCGGUCGC-GAGACUCUUGGAGGUAGAGCACUGUUUGAUUGAGGGGUCCAUCCCGGAUUACCAAUCUCAGAUAAACUCCGAAUGCCAACGAGUUAAGACCGGCAGUCAGACUG .((..((((((((..-..(((((((((.((.......(((...(((((.(..((((.....))))..)))))).))).......)).....)))).)))))..))))))).)..)).... ( -39.44) >sp_pneumo.rRNA 17812 120 + 2038623 GCUAGCGUCGACAUUAGAGAUUCUUGGAGGUAGAGCACUGUUUGGGUGAGGGGUCCAUCCCGGAUUACCAAUCUCAGAUAAACUCCGAAUGCCAAUGAAUUAUGGUCGGCAGUCAGACUG .((..(((((((....(.(((((((((.....(((...((((((((...(((((((.....))))).))...))))))))..)))......)))).))))).).)))))).)..)).... ( -33.50) >consensus GCUAGCGUCGAUGUU_AAGACUCUUGGAGGUAGAGCACUGUUUGGGUGAGGGGUCCAUCCCGGAUUACCAAUCUCAGAUAAACUCCGAAUGCCAACGAAUUAAAAUCGGCAGUCAGACUG .((..(((((((......(((((((((.....(((...((((((((...(((((((.....))))).))...))))))))..)))......)))).)))))...)))))).)..)).... (-35.75 = -32.62 + -3.12)


| Location | 580,763 – 580,883 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.97 |
| Mean single sequence MFE | -36.30 |
| Consensus MFE | -30.33 |
| Energy contribution | -30.20 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 580763 120 - 2809430 UCUGGGCUGUUUCCCUUUCGAACACGGACCUUAUCACCCAUGUUCUGACUCCCAAGUUAAAUUAAUUGGCAUUCGGAGUUUGUCUGAAUUCGGUAACCCGAGAGGGGCCCCUCGUCCAAA ..(((((.(..(((((((((.......(((...(((..((..((((((...((((..........))))...))))))..))..)))....)))....)))))))))..)...))))).. ( -33.80) >sp_agal.rRNA 19141 120 - 2160275 UCUGGGCUGUUUCCCUUUCGACUACGGAUCUUAGCACUCGCAGUCUGACUGCCGAUUAUAUCUCGUUGGCAUUCGGAGUUUAUCUGAGAUUGGUAAUCCGGGAUGGACCCCUCACCCAAA ..((((.((..(((.(..((..(((.((((((((.....((..(((((.(((((((........))))))).)))))))....)))))))).)))...))..).))).....)))))).. ( -38.80) >sp_mutans.rRNA 19721 120 - 2030929 UCUGGGCUGUUUCCCUUUCGACUACGGAUCUUAGCACUCGCAGUCUGACUGCCGGUCUUAACUCGUUGGCAUUCGGAGUUUAUCUGAGAUUGGUAAUCCGGGAUGGACCCCUCAAUCAAA ...(((((...((((....((.(((.((((((((.....((..(((((.((((((.(.......))))))).)))))))....)))))))).))).)).)))).)).))).......... ( -35.00) >sp_pneumo.rRNA 17852 120 - 2038623 UCUGGGCUGUUUCCCUUUCGACUACGGAUCUUAGCACUCGCAGUCUGACUGCCGACCAUAAUUCAUUGGCAUUCGGAGUUUAUCUGAGAUUGGUAAUCCGGGAUGGACCCCUCACCCAAA ..((((.((..(((.(..((..(((.((((((((.((((((((.....))))((((((........)))...)))))))....)))))))).)))...))..).))).....)))))).. ( -37.60) >consensus UCUGGGCUGUUUCCCUUUCGACUACGGAUCUUAGCACUCGCAGUCUGACUGCCGAUCAUAACUCAUUGGCAUUCGGAGUUUAUCUGAGAUUGGUAAUCCGGGAUGGACCCCUCACCCAAA ..((((..(..(((.(((((..(((.((((((((.....((..(((((.((((((..........)))))).)))))))....)))))))).)))...))))).)))..)....)))).. (-30.33 = -30.20 + -0.13) # Strand winner: forward (0.60)


| Location | 580,843 – 580,963 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.72 |
| Mean single sequence MFE | -29.54 |
| Consensus MFE | -24.25 |
| Energy contribution | -25.75 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 580843 120 - 2809430 UUCUAAGCCAACAUCCUAGUUGUCUGGGCAACGCCACAUCCUUUUCCACUUAACAUAUAUUUUGGGACCUUAGCUGGUGGUCUGGGCUGUUUCCCUUUCGAACACGGACCUUAUCACCCA .....(((.((..(((..(((((....))))).....................((.......)))))..)).)))((((((..((((((((((......))).)))).))).)))))).. ( -25.80) >sp_agal.rRNA 19221 120 - 2160275 UUCUAAGCUAACAUCCUAGUUGUCUGUGCAACCCCACAUCCUUUUCCACUUAACAUAUAUUUUGGGACCUUAGCUGGUGGUCUGGGCUGUUUCCCUUUCGACUACGGAUCUUAGCACUCG ......(((((.((((((((((..((((......)))).........................((((...(((((.(.....).)))))..))))...)))))).)))).)))))..... ( -30.20) >sp_mutans.rRNA 19801 120 - 2030929 UUCUAAGCUAACAUCCUAGUUGUCUGUGCAACCCCACAUCCUUUUCCACUUAACAAUUAUUUGGGGACCUUAGCUGGUGGUCUGGGCUGUUUCCCUUUCGACUACGGAUCUUAGCACUCG ......(((((.((((((((((.....(((.(((...........(((..((.....))..)))(((((.........)))))))).)))........)))))).)))).)))))..... ( -31.62) >sp_pneumo.rRNA 17932 120 - 2038623 UUCUAAGCUAACAUCCUAGUUGUCUGUGCAACCCCACAUCCUUUUCCACUUAACAAUUAUUUUGGGACCUUAGCUGGUGGUCUGGGCUGUUUCCCUUUCGACUACGGAUCUUAGCACUCG ......(((((.((((((((((.....(((.(((...................(((.....)))(((((.........)))))))).)))........)))))).)))).)))))..... ( -30.52) >consensus UUCUAAGCUAACAUCCUAGUUGUCUGUGCAACCCCACAUCCUUUUCCACUUAACAAAUAUUUUGGGACCUUAGCUGGUGGUCUGGGCUGUUUCCCUUUCGACUACGGAUCUUAGCACUCG ......(((((.((((((((((..((((......)))).........................((((...(((((.(.....).)))))..))))...)))))).)))).)))))..... (-24.25 = -25.75 + 1.50) # Strand winner: forward (0.98)

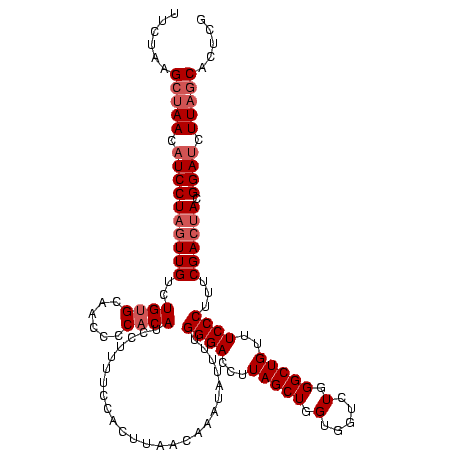
| Location | 580,923 – 581,041 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -36.45 |
| Consensus MFE | -34.79 |
| Energy contribution | -34.48 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.836754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 580923 118 + 2809430 GAUGUGGCGUUGCCCAGACAACUAGGAUGUUGGCUUAGAAGCAGCCA-UCAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACACUGCGCCGAAAAUGUACCGGGGCU-AAACAUA .((((......((((..(((.((((((((.(((((.......)))))-.))))....((((........)))).))))(((.(((......))))))...)))....)))).-..)))). ( -33.10) >sp_agal.rRNA 19301 120 + 2160275 GAUGUGGGGUUGCACAGACAACUAGGAUGUUAGCUUAGAAGCAGCUAUUCAUUCAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACCCUGCGCCGAAAAUGUACCGGGGCUAAAACAUA .(((((((((..(...(((.....(((((.(((((.......)))))..)))))....((((.((....)).)))).)))..)..)))))((.(((.........))).))....)))). ( -38.30) >sp_mutans.rRNA 19881 120 + 2030929 GAUGUGGGGUUGCACAGACAACUAGGAUGUUAGCUUAGAAGCAGCUAUUCAUUCAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACCCUGCGCCGAAAAUGUACCGGGGCUGAAACAAU ..((((((((..(...(((.....(((((.(((((.......)))))..)))))....((((.((....)).)))).)))..)..)))))((.(((.........))).))....))).. ( -37.20) >sp_pneumo.rRNA 18012 120 + 2038623 GAUGUGGGGUUGCACAGACAACUAGGAUGUUAGCUUAGAAGCAGCUAUUCAUUCAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACCCUGCGCCGAAAAUGUACCGGGGCUAAAACAAU ..((((((((..(...(((.....(((((.(((((.......)))))..)))))....((((.((....)).)))).)))..)..)))))((.(((.........))).))....))).. ( -37.20) >consensus GAUGUGGGGUUGCACAGACAACUAGGAUGUUAGCUUAGAAGCAGCUAUUCAUUCAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACCCUGCGCCGAAAAUGUACCGGGGCUAAAACAAA ..((((((((..(...(((.....(((((.(((((.......)))))..)))))....((((.((....)).)))).)))..)..)))))((.(((.........))).))....))).. (-34.79 = -34.48 + -0.31)

| Location | 580,923 – 581,041 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -32.25 |
| Consensus MFE | -30.48 |
| Energy contribution | -29.97 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 580923 118 - 2809430 UAUGUUU-AGCCCCGGUACAUUUUCGGCGCAGUGUCACUCGACUAGUGAGCUAUUACGCACUCUUUAAAUGA-UGGCUGCUUCUAAGCCAACAUCCUAGUUGUCUGGGCAACGCCACAUC .((((..-.((....))))))....((((...((((...(((((((.((((......))..........((.-(((((.......))))).)))))))))))....)))).))))..... ( -32.20) >sp_agal.rRNA 19301 120 - 2160275 UAUGUUUUAGCCCCGGUACAUUUUCGGCGCAGGGUCACUCGACUAGUGAGCUAUUACGCACUCUUUGAAUGAAUAGCUGCUUCUAAGCUAACAUCCUAGUUGUCUGUGCAACCCCACAUC .((((....((.((((.......)))).)).(((((((.(((((((.((((((((...((.....))....)))))).(((....))).....)))))))))...)))..)))).)))). ( -32.60) >sp_mutans.rRNA 19881 120 - 2030929 AUUGUUUCAGCCCCGGUACAUUUUCGGCGCAGGGUCACUCGACUAGUGAGCUAUUACGCACUCUUUGAAUGAAUAGCUGCUUCUAAGCUAACAUCCUAGUUGUCUGUGCAACCCCACAUC .........((.((((.......)))).)).(((((((.(((((((.((((((((...((.....))....)))))).(((....))).....)))))))))...)))..))))...... ( -32.10) >sp_pneumo.rRNA 18012 120 - 2038623 AUUGUUUUAGCCCCGGUACAUUUUCGGCGCAGGGUCACUCGACUAGUGAGCUAUUACGCACUCUUUGAAUGAAUAGCUGCUUCUAAGCUAACAUCCUAGUUGUCUGUGCAACCCCACAUC .........((.((((.......)))).)).(((((((.(((((((.((((((((...((.....))....)))))).(((....))).....)))))))))...)))..))))...... ( -32.10) >consensus AAUGUUUUAGCCCCGGUACAUUUUCGGCGCAGGGUCACUCGACUAGUGAGCUAUUACGCACUCUUUGAAUGAAUAGCUGCUUCUAAGCUAACAUCCUAGUUGUCUGUGCAACCCCACAUC .........((.((((.......)))).)).(((((((.(((((((.((((......))..............(((((.......)))))...)))))))))...)))..))))...... (-30.48 = -29.97 + -0.50) # Strand winner: forward (0.98)

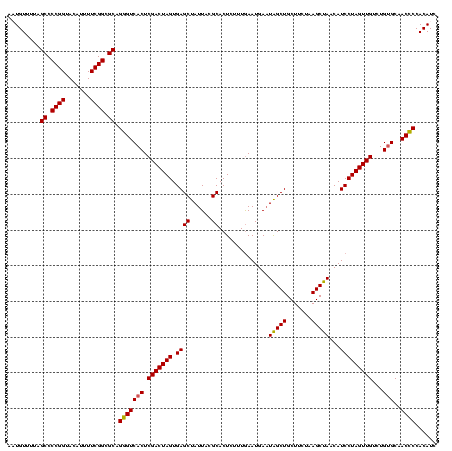
| Location | 580,963 – 581,080 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.99 |
| Mean single sequence MFE | -35.23 |
| Consensus MFE | -28.27 |
| Energy contribution | -28.21 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 580963 117 + 2809430 GCAGCCA-UCAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACACUGCGCCGAAAAUGUACCGGGGCU-AAACAUAUUACCGAAGCUGUGGAUUGUCCUUUGG-ACAAUGGUAGGA ((((...-(((((..(..((((.((....)).))))..)..)))))..))))(((.........(((.((((-..............)))).)))(((((((...))-)))))))).... ( -34.84) >sp_agal.rRNA 19341 120 + 2160275 GCAGCUAUUCAUUCAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACCCUGCGCCGAAAAUGUACCGGGGCUAAAACAUAUUACCGAAGCUGUGGAUGACACAAAAGUGUCAUGGUAGGA ((((....((((((.(..((((.((....)).))))..).))))))..))))(((.........(((.((((...............)))).)))(((((((....)))))))))).... ( -36.56) >sp_mutans.rRNA 19921 116 + 2030929 GCAGCUAUUCAUUCAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACCCUGCGCCGAAAAUGUACCGGGGCUGAAACAAUUUACCGAAGCUGUGGAUCCCUUAGGGG----AUGGUAGGA ((((....((((((.(..((((.((....)).))))..).))))))..))))(((.........(((.((((...............)))).)))(((((....)))----))))).... ( -35.76) >sp_pneumo.rRNA 18052 118 + 2038623 GCAGCUAUUCAUUCAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACCCUGCGCCGAAAAUGUACCGGGGCUAAAACAAUUUACCGAAGCUGUGGAUACCUUUAUAG--GUAUGGUAGGA ((((....((((((.(..((((.((....)).))))..).))))))..))))(((.........(((.((((...............)))).)))((((((....))--))))))).... ( -33.76) >consensus GCAGCUAUUCAUUCAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACCCUGCGCCGAAAAUGUACCGGGGCUAAAACAAAUUACCGAAGCUGUGGAUACCUCAAUAG____AUGGUAGGA ((((....((((((.(..((((.((....)).))))..).))))))..))))(((.........(((.((((...............)))).)))(((((........)))))))).... (-28.27 = -28.21 + -0.06)

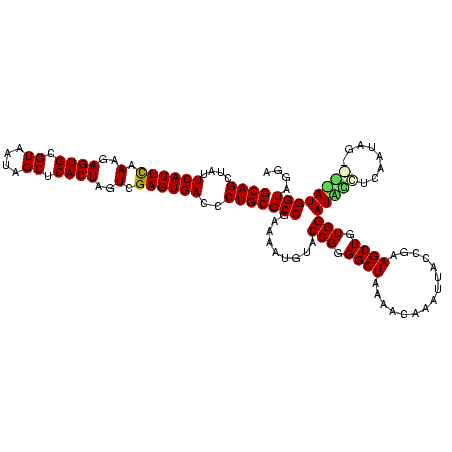
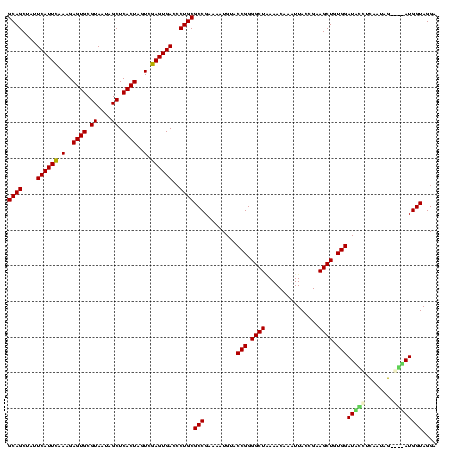
| Location | 581,041 – 581,160 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.01 |
| Mean single sequence MFE | -40.95 |
| Consensus MFE | -34.46 |
| Energy contribution | -33.65 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.47 |
| Mean z-score | -3.58 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.73 |
| SVM RNA-class probability | 0.999568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 581041 119 + 2809430 UUACCGAAGCUGUGGAUUGUCCUUUGG-ACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGUAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAA .((((..........(((((((...))-)))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))))))........... ( -37.90) >sp_agal.rRNA 19421 120 + 2160275 UUACCGAAGCUGUGGAUGACACAAAAGUGUCAUGGUAGGAGAGCGUUCUAUGUGUGAAGAAGGUGUACCGUGAGGAGCGCUGGAACGCAUAGAAGUGAGAAUGCCGGUAUGAGUAGCGCA .((((..........(((((((....)))))))((((.....((.((((((((((......(((((.((....)).)))))...)))))))))))).....))))))))........... ( -43.90) >sp_mutans.rRNA 20001 116 + 2030929 UUACCGAAGCUGUGGAUCCCUUAGGGG----AUGGUAGGAGAGCGUUCUAUGUGCGCAGAAGGUGUACCGCAAGGAGCGCUGGAGUGCAUAGAAGUGAGAAUGCCGGUAUGAGUAGCGUA .((((..........(((((....)))----))((((.....((.(((((((..(.(....(((((.((....)).))))).).)..))))))))).....))))))))........... ( -42.40) >sp_pneumo.rRNA 18132 118 + 2038623 UUACCGAAGCUGUGGAUACCUUUAUAG--GUAUGGUAGGAGAGCGUUCUAUGUGUGAUGAAGGUAUACCGUGAGGAGUGCUGGAACGCAUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAA .((((..........((((((....))--))))((((.....((.((((((((((......(((((.((....)).)))))...)))))))))))).....))))))))........... ( -39.60) >consensus UUACCGAAGCUGUGGAUACCUCAAUAG____AUGGUAGGAGAGCGUUCUAUGUGCGAAGAAGGUGUACCGUAAGGAGCGCUGGAACGCAUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAA .((((..........(((((........)))))((((.....((.((((((((((......(((((.((....)).)))))...)))))))))))).....))))))))........... (-34.46 = -33.65 + -0.81)

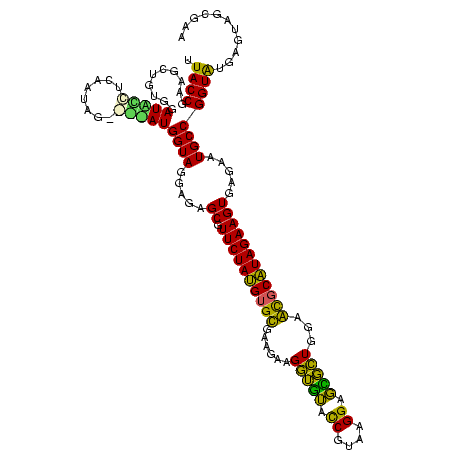
| Location | 581,080 – 581,200 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.08 |
| Mean single sequence MFE | -39.50 |
| Consensus MFE | -37.28 |
| Energy contribution | -36.65 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 581080 120 + 2809430 GAGCGUUCUAAGGGCGUUGAAGCAUGAUCGUAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGAUUGACUAAGGUUUC ..(((((((..(((((((...(((((.((....)).)))))..))))))).......)))))))(((((.((....(....).((((.......)))))))))))............... ( -38.90) >sp_agal.rRNA 19461 120 + 2160275 GAGCGUUCUAUGUGUGAAGAAGGUGUACCGUGAGGAGCGCUGGAACGCAUAGAAGUGAGAAUGCCGGUAUGAGUAGCGCAAGACAGGUGAGAAUCCUGUCCACCGUAAGACUAAGGUUUC ..((.((((((((((......(((((.((....)).)))))...))))))))))))..(((.(((......(((.(((...((((((.......))))))...)))...)))..)))))) ( -39.70) >sp_mutans.rRNA 20037 120 + 2030929 GAGCGUUCUAUGUGCGCAGAAGGUGUACCGCAAGGAGCGCUGGAGUGCAUAGAAGUGAGAAUGCCGGUAUGAGUAGCGUAAGACAGGUGAGAAUCCUGUCCACCGUAAGACUAAGGAUUC (.(((((((.(((((((....(((((.((....)).)))))...)))))))......))))))))......(((.(((...((((((.......))))))...)))...)))........ ( -41.20) >sp_pneumo.rRNA 18170 120 + 2038623 GAGCGUUCUAUGUGUGAUGAAGGUAUACCGUGAGGAGUGCUGGAACGCAUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGACAGGUGAGAAUCCUGUCCACCGUAAGACUAAGGUUUC ..((.((((((((((......(((((.((....)).)))))...))))))))))))..(((.(((......(((.(((...((((((.......))))))...)))...)))..)))))) ( -38.20) >consensus GAGCGUUCUAUGUGCGAAGAAGGUGUACCGUAAGGAGCGCUGGAACGCAUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGACAGGUGAGAAUCCUGUCCACCGUAAGACUAAGGUUUC ..((.((((((((((......(((((.((....)).)))))...))))))))))))..(((.(((......(((.(((...((((((.......))))))...)))...)))..)))))) (-37.28 = -36.65 + -0.63)

| Location | 581,120 – 581,240 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.75 |
| Mean single sequence MFE | -34.33 |
| Consensus MFE | -33.10 |
| Energy contribution | -32.60 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 581120 120 - 2809430 UUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCA ...(((.((((((((.((((((.(((......))))))))).....))))))..((((((((((((.......))))))....(.....).))))))...............).).))). ( -34.50) >sp_agal.rRNA 19501 120 - 2160275 UUAGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAACCUUAGUCUUACGGUGGACAGGAUUCUCACCUGUCUUGCGCUACUCAUACCGGCAUUCUCACUUCUAUGCGUUCCA ...((..(.((((((.((((((.(((......))))))))).....))))))...(((((((((((.......)))))))............))))((((..........)))))..)). ( -34.50) >sp_mutans.rRNA 20077 120 - 2030929 UUAGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAUCCUUAGUCUUACGGUGGACAGGAUUCUCACCUGUCUUACGCUACUCAUACCGGCAUUCUCACUUCUAUGCACUCCA ...(((...((((((.((((((.(((......))))))))).....))))))...((..(((((((.......)))))))..))........))).((((..........))))...... ( -33.70) >sp_pneumo.rRNA 18210 120 - 2038623 UUAGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAACCUUAGUCUUACGGUGGACAGGAUUCUCACCUGUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAUGCGUUCCA ...(((...((((((.((((((.(((......))))))))).....))))))...(((.(((((((.......))))))).)))........))).((((..........))))...... ( -34.60) >consensus UUAGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAACCUUAGUCUUACGGUGGACAGGAUUCUCACCUGUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAUGCGCUCCA ...((....((((((.((((((.(((......))))))))).....))))))...(((((((((((.......)))))))............))))((((..........))))...)). (-33.10 = -32.60 + -0.50) # Strand winner: forward (1.00)

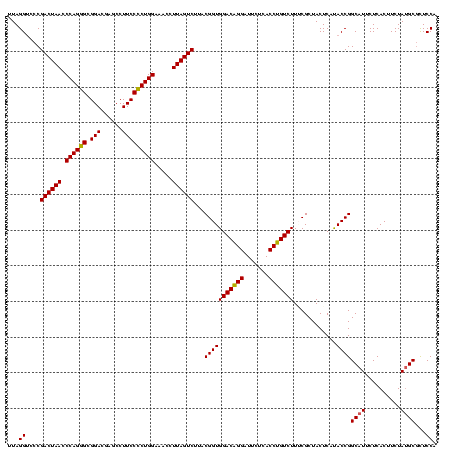
| Location | 581,160 – 581,280 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -44.08 |
| Consensus MFE | -44.53 |
| Energy contribution | -42.65 |
| Covariance contribution | -1.88 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.80 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 581160 120 + 2809430 AGACGGGUGAGAAUCCCGUCCACCGAUUGACUAAGGUUUCCAGAGGAAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGACAGGCGUAGGCGAUGGAUAACAGGUUGAU .((((((.......))))))..(((((((((........((((((...((.....)).)))))))))))))))((((...(((.(.(((....))).).)))...))))........... ( -46.10) >sp_agal.rRNA 19541 120 + 2160275 AGACAGGUGAGAAUCCUGUCCACCGUAAGACUAAGGUUUCCAGGGGAAGGCUCGUCCGCCCUGGGUUAGUCGGGACCUAAGGAGAGACCGAAAGGUGUAUCCGAUGGACAACAGGUUGAU .((((((.......)))))).(((....((((((....(((((((...((.....)).)))))))))))))....((...(((.(.(((....))).).)))...))......))).... ( -44.00) >sp_mutans.rRNA 20117 120 + 2030929 AGACAGGUGAGAAUCCUGUCCACCGUAAGACUAAGGAUUCCAGGGGAAGGCUCGUCCGCCCUGGGUUAGUCGGGACCUAAGGAGAGACCGAUAGGUGUAUCCGAUGGGCAACAGGUUGAU .((((((.......)))))).(((....((((((....(((((((...((.....)).)))))))))))))..(.((((.(((.(.(((....))).).)))..)))))....))).... ( -42.20) >sp_pneumo.rRNA 18250 120 + 2038623 AGACAGGUGAGAAUCCUGUCCACCGUAAGACUAAGGUUUCCAGGGGAAGGCUCGUCCGCCCUGGGUUAGUCGGGACCUAAGGAGAGACCGAAAGGUGUAUCCGAUGGACAACAGGUUGAU .((((((.......)))))).(((....((((((....(((((((...((.....)).)))))))))))))....((...(((.(.(((....))).).)))...))......))).... ( -44.00) >consensus AGACAGGUGAGAAUCCUGUCCACCGUAAGACUAAGGUUUCCAGGGGAAGGCUCGUCCGCCCUGGGUUAGUCGGGACCUAAGGAGAGACCGAAAGGUGUAUCCGAUGGACAACAGGUUGAU .((((((.......)))))).(((....((((((....(((((((...((.....)).)))))))))))))....((...(((.(.(((....))).).)))...))......))).... (-44.53 = -42.65 + -1.88)

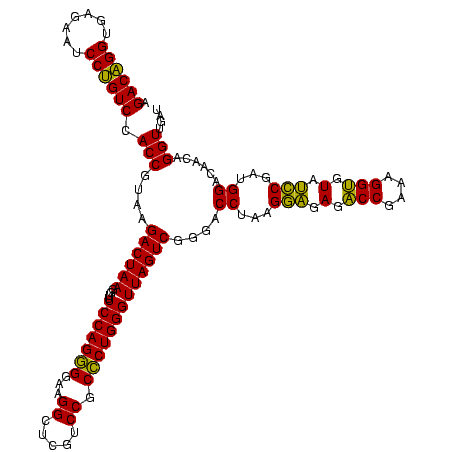
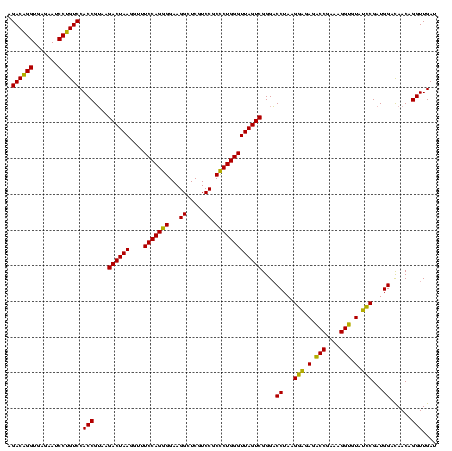
| Location | 581,160 – 581,280 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -37.28 |
| Consensus MFE | -35.78 |
| Energy contribution | -34.53 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 581160 120 - 2809430 AUCAACCUGUUAUCCAUCGCCUACGCCUGUCGGCCUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCU ....(((.(((.(((...((....(((....)))....))...)))...((((((.((((((.(((......))))))))).....))))))))).)))(((((((.......))))))) ( -36.60) >sp_agal.rRNA 19541 120 - 2160275 AUCAACCUGUUGUCCAUCGGAUACACCUUUCGGUCUCUCCUUAGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAACCUUAGUCUUACGGUGGACAGGAUUCUCACCUGUCU ..........(((((((((.....((((...((.....))..))))...((((((.((((((.(((......))))))))).....))))))...)))))))))((((........)))) ( -37.30) >sp_mutans.rRNA 20117 120 - 2030929 AUCAACCUGUUGCCCAUCGGAUACACCUAUCGGUCUCUCCUUAGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAUCCUUAGUCUUACGGUGGACAGGAUUCUCACCUGUCU .....(((((...((((((.....(((((..((.....)).)))))...((((((.((((((.(((......))))))))).....))))))...))))))))))).............. ( -37.90) >sp_pneumo.rRNA 18250 120 - 2038623 AUCAACCUGUUGUCCAUCGGAUACACCUUUCGGUCUCUCCUUAGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAACCUUAGUCUUACGGUGGACAGGAUUCUCACCUGUCU ..........(((((((((.....((((...((.....))..))))...((((((.((((((.(((......))))))))).....))))))...)))))))))((((........)))) ( -37.30) >consensus AUCAACCUGUUGUCCAUCGGAUACACCUUUCGGUCUCUCCUUAGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAACCUUAGUCUUACGGUGGACAGGAUUCUCACCUGUCU ....(((......((...(((.(.(((....))).).)))...))....((((((.((((((.(((......))))))))).....))))))....)))(((((((.......))))))) (-35.78 = -34.53 + -1.25) # Strand winner: forward (1.00)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Fri Apr 7 15:50:52 2006