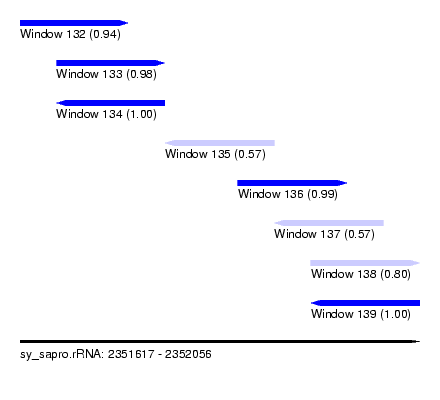
| Sequence ID | sy_sapro.rRNA |
|---|---|
| Location | 2,351,617 – 2,352,056 |
| Length | 439 |
| Max. P | 0.997464 |

| Location | 2,351,617 – 2,351,736 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.42 |
| Mean single sequence MFE | -28.37 |
| Consensus MFE | -26.31 |
| Energy contribution | -25.53 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2351617 119 + 2516583 ACAAUUCUAGCUAGAGGCUUUUCUUGGCAGUGUGAAAUCAACGACUCGAGGAAACAAUUUCCUCUCCCCAUCACAGCUCAAUCUUAA-GAGUGCCGGAUUUGCCUAACACUCAAUCUCAC ..............((((...(((.(((..(((((..((...))...((((((.....))))))......)))))((((........-))))))))))...))))............... ( -27.00) >sy_haemo.rRNA 2540506 119 - 2685023 UAUUAUCUAUCUAGAGGCUUUUCUCGGCAGUGUGAAAUCAACGACUCGAGGAAACAAUUUCCUCUCCCCAUCACAGCUCAAUCUUAA-GAGUGCCGGAUUUGCCUAACACUCAAUCUCAC ..............((((...((.((((..(((((..((...))...((((((.....))))))......)))))((((........-))))))))))...))))............... ( -29.20) >sy_aureus.rRNA 2276724 120 - 2809430 UAUUUUCUAUCUAGAGGCUUUUCUCGGCAGUGUGAAAUCAACGACUCGAAGACACAAUGUCUUCUCCCCAUCACAGCUCAGCCUUAACGAGUACCGGAUUUGCCUAAUACUCAGCCUUAC .............((((((......((((((((((..((...))...((((((.....))))))......))))).))..))).....(((((..((.....))...))))))))))).. ( -28.90) >consensus UAUUUUCUAUCUAGAGGCUUUUCUCGGCAGUGUGAAAUCAACGACUCGAGGAAACAAUUUCCUCUCCCCAUCACAGCUCAAUCUUAA_GAGUGCCGGAUUUGCCUAACACUCAAUCUCAC ..............((((...((.((((..(((((..((...))...((((((.....))))))......)))))((((.........))))))))))...))))............... (-26.31 = -25.53 + -0.77)

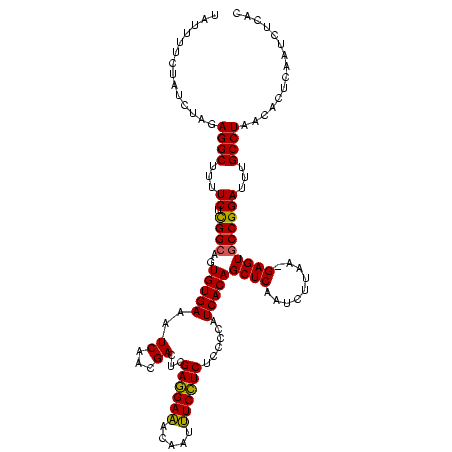
| Location | 2,351,657 – 2,351,776 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.64 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -24.79 |
| Energy contribution | -22.80 |
| Covariance contribution | -1.99 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.96 |
| Structure conservation index | 1.03 |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.981416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2351657 119 + 2516583 ACGACUCGAGGAAACAAUUUCCUCUCCCCAUCACAGCUCAAUCUUAA-GAGUGCCGGAUUUGCCUAACACUCAAUCUCACUGCUUGGACGUACAAUCCAAUCGUACGCUUCGCCUAUCCU .......((((((.....)))))).......................-(((((..((.....))...))))).........((..((.(((((.........)))))))..))....... ( -22.60) >sy_haemo.rRNA 2540546 119 - 2685023 ACGACUCGAGGAAACAAUUUCCUCUCCCCAUCACAGCUCAAUCUUAA-GAGUGCCGGAUUUGCCUAACACUCAAUCUCACUGCUUAGACGUACAAUCCAAUCGUACGCUUCGCCUAUCCU .......((((((.....)))))).......................-(((((..((.....))...))))).........((..((.(((((.........)))))))..))....... ( -22.60) >sy_aureus.rRNA 2276764 120 - 2809430 ACGACUCGAAGACACAAUGUCUUCUCCCCAUCACAGCUCAGCCUUAACGAGUACCGGAUUUGCCUAAUACUCAGCCUUACUGCUUAGACGUGCAAUCCAAUCGCACGCUUCGCCUAUCCU ...((((((((((.....))))))...........((...))......))))...((((..((......((.(((......))).)).(((((.........)))))....))..)))). ( -27.10) >consensus ACGACUCGAGGAAACAAUUUCCUCUCCCCAUCACAGCUCAAUCUUAA_GAGUGCCGGAUUUGCCUAACACUCAAUCUCACUGCUUAGACGUACAAUCCAAUCGUACGCUUCGCCUAUCCU .......((((((.....))))))........................(((((..((.....))...))))).........((..((.(((((.........)))))))..))....... (-24.79 = -22.80 + -1.99)

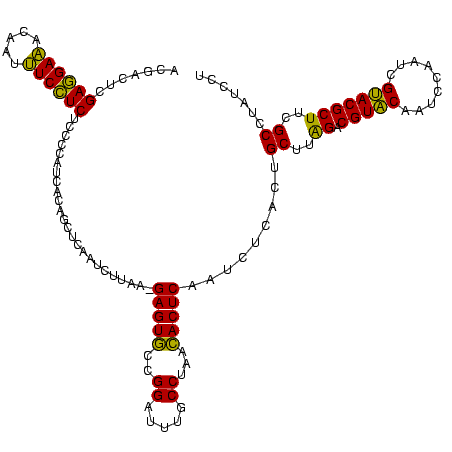
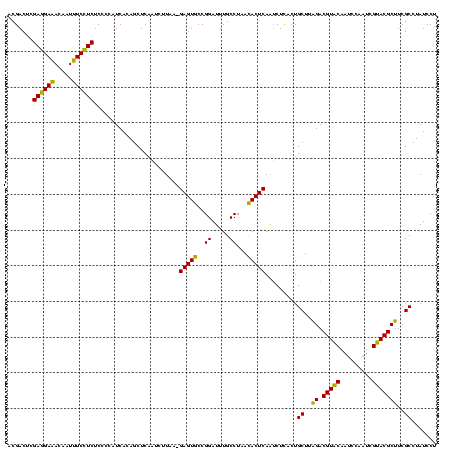
| Location | 2,351,657 – 2,351,776 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.64 |
| Mean single sequence MFE | -38.67 |
| Consensus MFE | -41.28 |
| Energy contribution | -38.40 |
| Covariance contribution | -2.88 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.33 |
| Structure conservation index | 1.07 |
| SVM decision value | 2.86 |
| SVM RNA-class probability | 0.997464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2351657 119 - 2516583 AGGAUAGGCGAAGCGUACGAUUGGAUUGUACGUCCAAGCAGUGAGAUUGAGUGUUAGGCAAAUCCGGCACUC-UUAAGAUUGAGCUGUGAUGGGGAGAGGAAAUUGUUUCCUCGAGUCGU ......(((...(((((((((...)))))))))(((.(((((..((((((((((..((....))..))))))-....))))..)))))..)))...((((((.....))))))..))).. ( -39.20) >sy_haemo.rRNA 2540546 119 + 2685023 AGGAUAGGCGAAGCGUACGAUUGGAUUGUACGUCUAAGCAGUGAGAUUGAGUGUUAGGCAAAUCCGGCACUC-UUAAGAUUGAGCUGUGAUGGGGAGAGGAAAUUGUUUCCUCGAGUCGU ......(((...(((((((((...)))))))))(((.(((((..((((((((((..((....))..))))))-....))))..)))))..)))...((((((.....))))))..))).. ( -36.90) >sy_aureus.rRNA 2276764 120 + 2809430 AGGAUAGGCGAAGCGUGCGAUUGGAUUGCACGUCUAAGCAGUAAGGCUGAGUAUUAGGCAAAUCCGGUACUCGUUAAGGCUGAGCUGUGAUGGGGAGAAGACAUUGUGUCUUCGAGUCGU ......(((...(((((((((...)))))))))(((.(((((..((((((((((..((....))..)))))).....))))..)))))..)))...((((((.....))))))..))).. ( -39.90) >consensus AGGAUAGGCGAAGCGUACGAUUGGAUUGUACGUCUAAGCAGUGAGAUUGAGUGUUAGGCAAAUCCGGCACUC_UUAAGAUUGAGCUGUGAUGGGGAGAGGAAAUUGUUUCCUCGAGUCGU ......(((...(((((((((...)))))))))(((.(((((..((((((((((..((....))..)))))).....))))..)))))..)))...((((((.....))))))..))).. (-41.28 = -38.40 + -2.88) # Strand winner: reverse (1.00)



| Location | 2,351,776 – 2,351,896 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -44.73 |
| Consensus MFE | -41.97 |
| Energy contribution | -42.30 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2351776 120 - 2516583 AAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGAUAGGCGUAGGCGAUGGAUAACAGGUUGAUAUUCCUGUACCACCAUUAUUCGUUUUAAGCGAUGGGGGGACGCAGU ...((((((((.(((..(((.....((((((...(((.(.(((....))).).)))...)))).(((((........))))).))))).....((((.....)))).))).))))).))) ( -42.80) >sy_haemo.rRNA 2540665 120 + 2685023 AAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGACAGGCGUAGGCGAUGGAUAACAGGUUGAUAUUCCUGUACCACCUAGCAUCGUUUUAAUCGAUGGGGGGACGCAGU ...((((((((.(((...(((((..((((((...(((.(.(((....))).).)))...)))).(((((........))))).))..)))))((((.......))))))).))))).))) ( -47.20) >sy_aureus.rRNA 2276884 120 + 2809430 AAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGACAGGCGUAGGCGAUGGAUAACAGGUUGAUAUUCCUGUACCACCUAUAAUCGUUUUAAUCGAUGGGGGGACGCAGU ...((((((((((.(((.((..(((((..(((......))))))))..)).)))))..(((...(((((........))))).)))(((...((((.......)))).)))))))).))) ( -44.20) >consensus AAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGACAGGCGUAGGCGAUGGAUAACAGGUUGAUAUUCCUGUACCACCUAUAAUCGUUUUAAUCGAUGGGGGGACGCAGU ...((((((((.((((.((((((.(((((((...(((.(.(((....))).).)))...)))).(((((........)))))...........)))..)))))).).))).))))).))) (-41.97 = -42.30 + 0.33) # Strand winner: reverse (1.00)

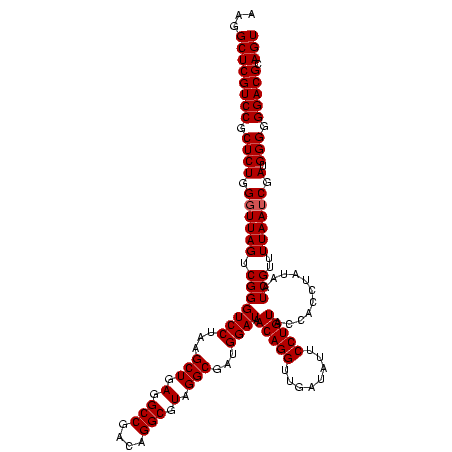
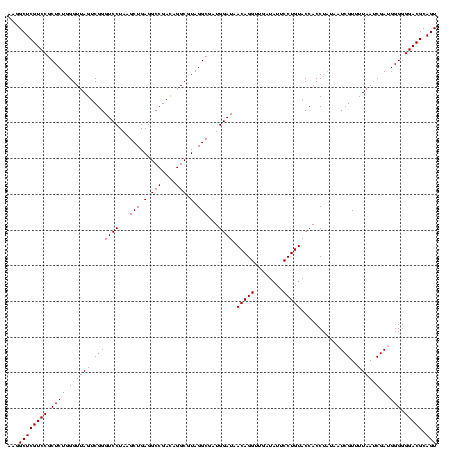
| Location | 2,351,856 – 2,351,976 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -39.50 |
| Consensus MFE | -39.50 |
| Energy contribution | -39.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.09 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2351856 120 + 2516583 CUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGC ....((((((((...((((((.((((((.(((......))))))))).....))))))..((((((((((((.......))))))....(.....).)))))).........)))))))) ( -39.50) >sy_haemo.rRNA 2540745 120 - 2685023 CUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGC ....((((((((...((((((.((((((.(((......))))))))).....))))))..((((((((((((.......))))))....(.....).)))))).........)))))))) ( -39.50) >sy_aureus.rRNA 2276964 120 - 2809430 CUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGC ....((((((((...((((((.((((((.(((......))))))))).....))))))..((((((((((((.......))))))....(.....).)))))).........)))))))) ( -39.50) >consensus CUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGC ....((((((((...((((((.((((((.(((......))))))))).....))))))..((((((((((((.......))))))....(.....).)))))).........)))))))) (-39.50 = -39.50 + 0.00)

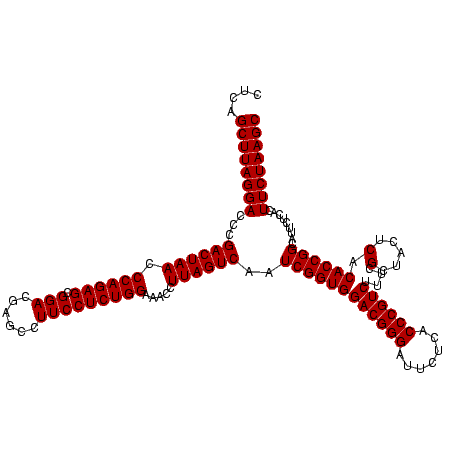
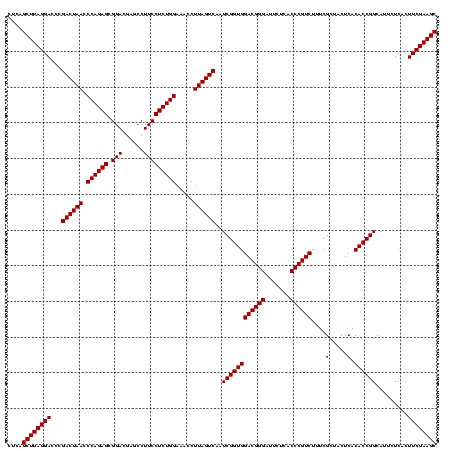
| Location | 2,351,896 – 2,352,016 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -37.97 |
| Consensus MFE | -37.97 |
| Energy contribution | -37.97 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.38 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2351896 120 - 2516583 UCUAAGGGCGUUGAAGCAUGAUCGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGAUUGACUAAGGUUUCCAGAGG ((((((.((......(((((.((....)).)))))...)).))))))..((.(((.(((...((.(((..((...((((((.......))))))...))))).))...)))))))).... ( -38.30) >sy_haemo.rRNA 2540785 120 + 2685023 UCUAAGGGCGUUGAAGCAUGAUCGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGAUUGACUAAGGUUUCCAGAGG ((((((.((......(((((.((....)).)))))...)).))))))..((.(((.(((...((.(((..((...((((((.......))))))...))))).))...)))))))).... ( -38.30) >sy_aureus.rRNA 2277004 120 + 2809430 UCUAAGGGCGUUGAAGCAUGAUCGUAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGAUUGACUAAGGUUUCCAGAGG ((((((.((......(((((.((....)).)))))...)).))))))..((.(((.(((...((.(((..((...((((((.......))))))...))))).))...)))))))).... ( -37.30) >consensus UCUAAGGGCGUUGAAGCAUGAUCGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGAUUGACUAAGGUUUCCAGAGG ((((((.((......(((((.((....)).)))))...)).))))))..((.(((.(((...((.(((..((...((((((.......))))))...))))).))...)))))))).... (-37.97 = -37.97 + -0.00) # Strand winner: reverse (1.00)

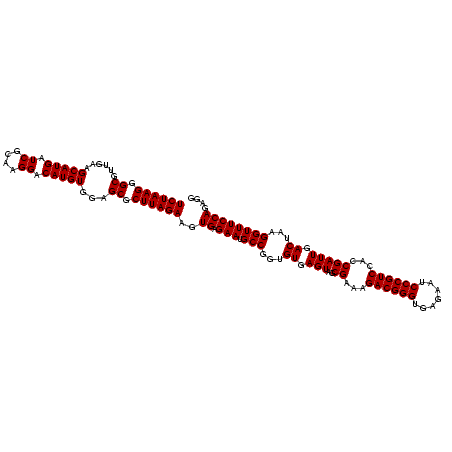

| Location | 2,351,936 – 2,352,056 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.77 |
| Mean single sequence MFE | -20.41 |
| Consensus MFE | -21.44 |
| Energy contribution | -21.44 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 1.05 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2351936 120 + 2516583 CCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUGCGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCCUUACGGACAAUCCACAGCUU .........(((.........((.........(((((((.((....((((((.....)).)))).......)).)))))))..........))(((((((....)))))))....))).. ( -21.41) >sy_haemo.rRNA 2540825 119 - 2685023 CCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUGCGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCCU-ACGGACAAUCCACAGCUU .........(((.........((.........(((((((.((....((((((.....)).)))).......)).)))))))..........))(((((((.-..)))))))....))).. ( -19.91) >sy_aureus.rRNA 2277044 119 - 2809430 CCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUACGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCCA-AAGGACAAUCCACAGCUU .........(((.........((.........(((((((.((....((((((.....)).)))).......)).)))))))..........))(((((((.-..)))))))....))).. ( -19.91) >consensus CCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUGCGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCCU_ACGGACAAUCCACAGCUU .........(((.........((.........(((((((.((....((((((.....)).)))).......)).)))))))..........))(((((((....)))))))....))).. (-21.44 = -21.44 + 0.00)

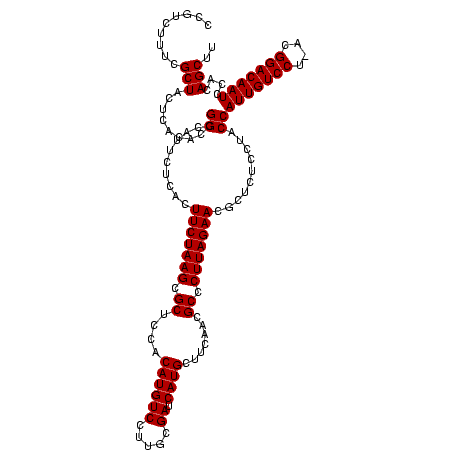
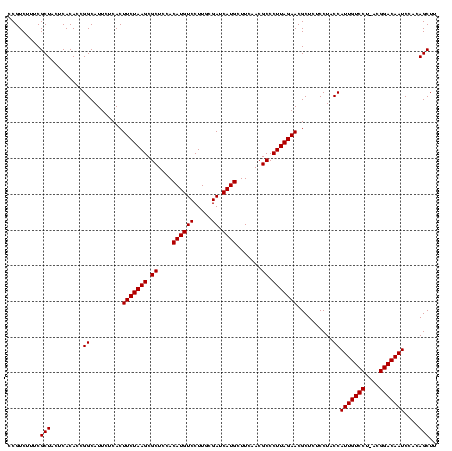
| Location | 2,351,936 – 2,352,056 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.77 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -41.37 |
| Energy contribution | -41.37 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.60 |
| Structure conservation index | 1.04 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2351936 120 - 2516583 AAGCUGUGGAUUGUCCGUAAGGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGG ..((((...(((((((....)))))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))........))))......... ( -43.10) >sy_haemo.rRNA 2540825 119 + 2685023 AAGCUGUGGAUUGUCCGU-AGGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGG ..((((...(((((((..-.)))))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))........))))......... ( -38.80) >sy_aureus.rRNA 2277044 119 + 2809430 AAGCUGUGGAUUGUCCUU-UGGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGUAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGG ..((((...(((((((..-.)))))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))........))))......... ( -37.80) >consensus AAGCUGUGGAUUGUCCGU_AGGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGG ..((((...(((((((....)))))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))........))))......... (-41.37 = -41.37 + 0.00) # Strand winner: reverse (1.00)

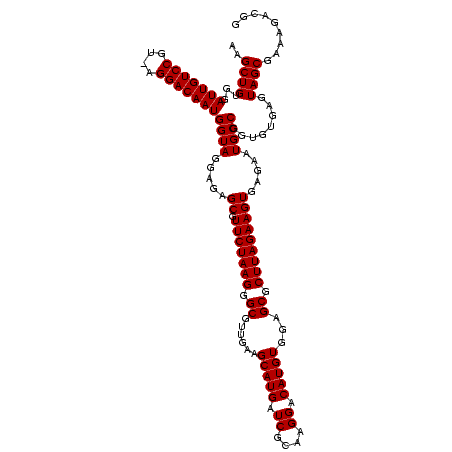
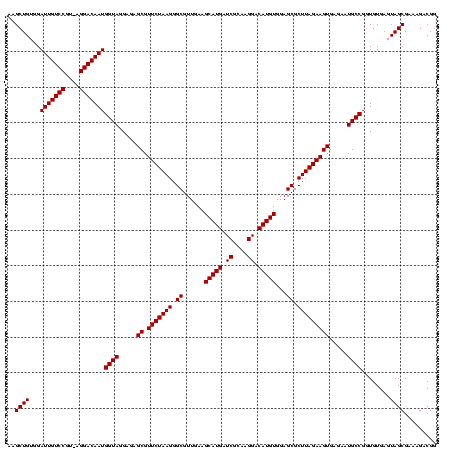
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Fri Apr 7 15:52:24 2006