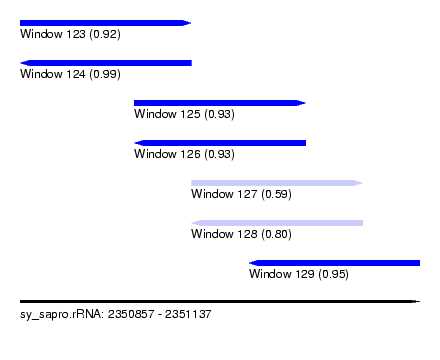
| Sequence ID | sy_sapro.rRNA |
|---|---|
| Location | 2,350,857 – 2,351,137 |
| Length | 280 |
| Max. P | 0.993672 |

| Location | 2,350,857 – 2,350,977 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -34.36 |
| Energy contribution | -33.70 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.88 |
| Structure conservation index | 1.02 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2350857 120 + 2516583 CUGCUCGACUUGUAAGUCUCGCAGUCAAGCUUCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGAAACCUUUGAGCGCCUCCGUUACUCUUUAGGAGGCGACCGCCC ((((..((((....))))..))))(((((.(((((((((......))......(((((.........))))))))))))...))))).(((((((...........)))))))....... ( -31.90) >sy_haemo.rRNA 2539746 120 - 2685023 CUGCUCGACUUGUAGGUCUCGCAGUCAAGCUCCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCC ((((..((((....))))..))))(((((.(((((((((......))......(((((.........))))))))))))...))))).(((((((...........)))))))....... ( -34.60) >sy_aureus.rRNA 2275964 120 - 2809430 CUGCUCGACUUGUAGGUCUCGCAGUCAAGCUCCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCC ((((..((((....))))..))))(((((.(((((((((......))......(((((.........))))))))))))...))))).(((((((...........)))))))....... ( -34.60) >consensus CUGCUCGACUUGUAGGUCUCGCAGUCAAGCUCCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCC ((((..((((....))))..))))(((((.(((((((((......))......(((((.........))))))))))))...))))).(((((((...........)))))))....... (-34.36 = -33.70 + -0.66)


| Location | 2,350,857 – 2,350,977 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -43.50 |
| Consensus MFE | -43.78 |
| Energy contribution | -43.33 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.04 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2350857 120 - 2516583 GGGCGGUCGCCUCCUAAAGAGUAACGGAGGCGCUCAAAGGUUUCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGAAGCUUGACUGCGAGACUUACAAGUCGAGCAG ..((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((((....)))).)))))))))))))))..((((....))))...... ( -42.70) >sy_haemo.rRNA 2539746 120 + 2685023 GGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAG ..((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((((....)))).)))))))))))))))..(((......)))...... ( -43.90) >sy_aureus.rRNA 2275964 120 + 2809430 GGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAG ..((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((((....)))).)))))))))))))))..(((......)))...... ( -43.90) >consensus GGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAG ..((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((((....)))).)))))))))))))))..(((......)))...... (-43.78 = -43.33 + -0.44) # Strand winner: reverse (1.00)

| Location | 2,350,937 – 2,351,057 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -42.27 |
| Consensus MFE | -41.11 |
| Energy contribution | -40.67 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2350937 120 + 2516583 CCUUUGAGCGCCUCCGUUACUCUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCAUCUGACACUGUCUCCCACCAUGAUAAAUGGUGCGGGUUAGAAAUCCAACACAGCGAGGGUAGUAU .....(..(((((((...........)))))))..)...........(((((((.((((((.(((.....((((((.....)))))))))))))))..(((......).))))))))).. ( -40.40) >sy_haemo.rRNA 2539826 120 - 2685023 CCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAGCUAGGGUAGUAU .....(..(((((((...........)))))))..)...........(((((((..(((((.(((.....((((((.....))))))))))))))..(((.......))).))))))).. ( -43.20) >sy_aureus.rRNA 2276044 120 - 2809430 CCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAGCUAGGGUAGUAU .....(..(((((((...........)))))))..)...........(((((((..(((((.(((.....((((((.....))))))))))))))..(((.......))).))))))).. ( -43.20) >consensus CCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAGCUAGGGUAGUAU .....(..(((((((...........)))))))..)...........(((((((..(((.........((((((((.....)))))).))(((.(....).))).)))...))))))).. (-41.11 = -40.67 + -0.44)

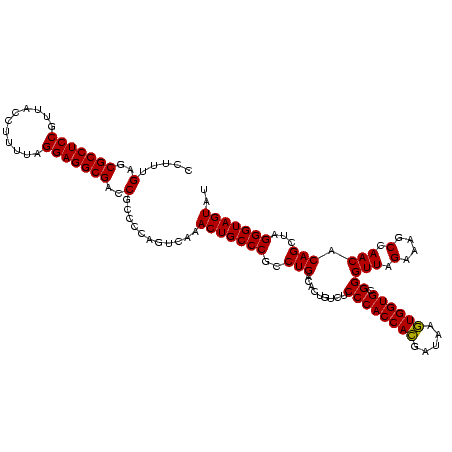
| Location | 2,350,937 – 2,351,057 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -47.23 |
| Consensus MFE | -47.68 |
| Energy contribution | -47.13 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.92 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2350937 120 - 2516583 AUACUACCCUCGCUGUGUUGGAUUUCUAACCCGCACCAUUUAUCAUGGUGGGAGACAGUGUCAGAUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGAGUAACGGAGGCGCUCAAAGG ..((..((((.((((((((((....)))))((.((((((.....))))))))..)))))(((((((.....))))))).))))..))(((((((...........)))))))........ ( -44.50) >sy_haemo.rRNA 2539826 120 + 2685023 AUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGGCGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGG ..((..(((((((((((((((....)))))((.((((((.....))))))))..)))))(((((((.....))))))))))))..))(((((((...........)))))))........ ( -48.60) >sy_aureus.rRNA 2276044 120 + 2809430 AUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGGCGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGG ..((..(((((((((((((((....)))))((.((((((.....))))))))..)))))(((((((.....))))))))))))..))(((((((...........)))))))........ ( -48.60) >consensus AUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGGCGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGG ..((..(((((((((((((((....)))))((.((((((.....))))))))..)))))(((((((.....))))))))))))..))(((((((...........)))))))........ (-47.68 = -47.13 + -0.55) # Strand winner: reverse (1.00)


| Location | 2,350,977 – 2,351,097 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -33.21 |
| Energy contribution | -32.77 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
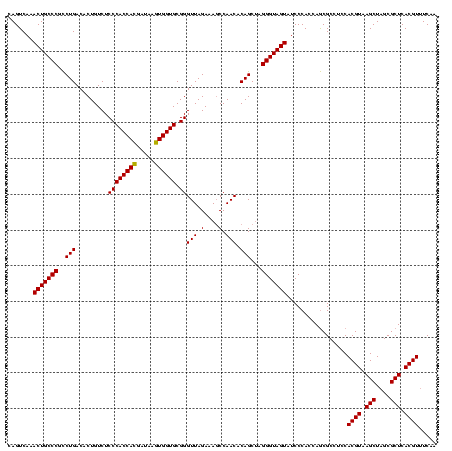
>sy_sapro.rRNA 2350977 120 + 2516583 CAGUCAAACUGCCCAUCUGACACUGUCUCCCACCAUGAUAAAUGGUGCGGGUUAGAAAUCCAACACAGCGAGGGUAGUAUCCCACCAACGCCUCCACGUAAGCUAGCGCUCACGUAUCUC .......(((((((.((((((.(((.....((((((.....)))))))))))))))..(((......).))))))))).................((((.(((....))).))))..... ( -32.90) >sy_haemo.rRNA 2539866 120 - 2685023 CAGUCAAACUGCCCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAGCGCUCACGUUUCAA .......(((((((..(((((.(((.....((((((.....))))))))))))))..(((.......))).)))))))........(((((.....(....)...))))).......... ( -35.30) >sy_aureus.rRNA 2276084 120 - 2809430 CAGUCAAACUGCCCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAGCGCUCACGUUUCAA .......(((((((..(((((.(((.....((((((.....))))))))))))))..(((.......))).)))))))........(((((.....(....)...))))).......... ( -35.30) >consensus CAGUCAAACUGCCCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAGCGCUCACGUUUCAA .......(((((((..(((.........((((((((.....)))))).))(((.(....).))).)))...))))))).................((((.(((....))).))))..... (-33.21 = -32.77 + -0.44)

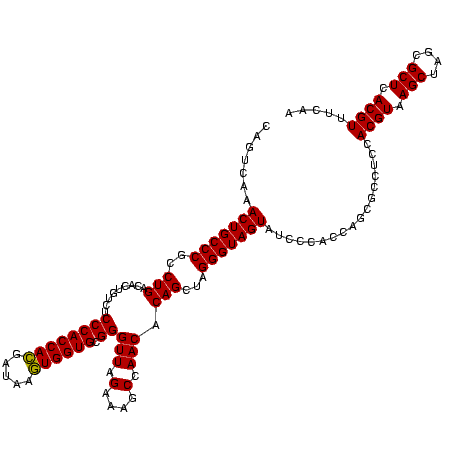
| Location | 2,350,977 – 2,351,097 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -46.20 |
| Consensus MFE | -45.22 |
| Energy contribution | -44.33 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2350977 120 - 2516583 GAGAUACGUGAGCGCUAGCUUACGUGGAGGCGUUGGUGGGAUACUACCCUCGCUGUGUUGGAUUUCUAACCCGCACCAUUUAUCAUGGUGGGAGACAGUGUCAGAUGGGCAGUUUGACUG (((.(((((((((....)))))))))........(((((....))))))))((((((((((....)))))((.((((((.....))))))))..)))))(((((((.....))))))).. ( -46.80) >sy_haemo.rRNA 2539866 120 + 2685023 UUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGGCGGGCAGUUUGACUG .....((((((((....)))))))).((.(((((((.((.......))))))).)).)).(((.(((...((.((((((.....))))))))))).)))(((((((.....))))))).. ( -45.90) >sy_aureus.rRNA 2276084 120 + 2809430 UUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGGCGGGCAGUUUGACUG .....((((((((....)))))))).((.(((((((.((.......))))))).)).)).(((.(((...((.((((((.....))))))))))).)))(((((((.....))))))).. ( -45.90) >consensus UUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGGCGGGCAGUUUGACUG .....((((((((....)))))))).........(((((....)))))...((((((((((....)))))((.((((((.....))))))))..)))))(((((((.....))))))).. (-45.22 = -44.33 + -0.88) # Strand winner: reverse (1.00)

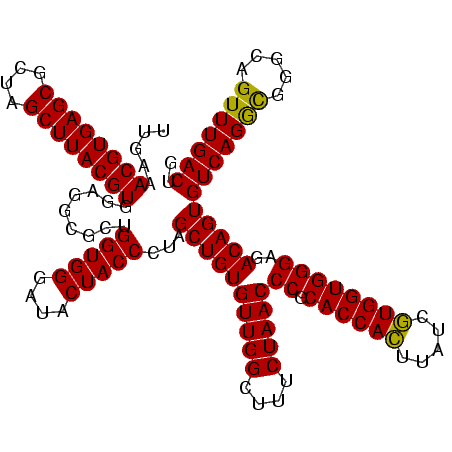
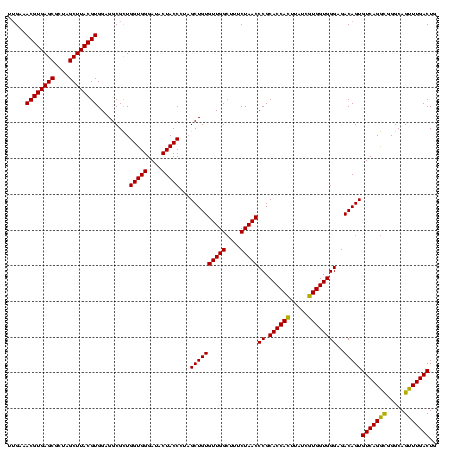
| Location | 2,351,017 – 2,351,137 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -46.90 |
| Consensus MFE | -43.63 |
| Energy contribution | -43.97 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2351017 120 - 2516583 UUGAAAUUCGGCACAGCUUGUACAGGAUAGGUAGGAGCCUGAGAUACGUGAGCGCUAGCUUACGUGGAGGCGUUGGUGGGAUACUACCCUCGCUGUGUUGGAUUUCUAACCCGCACCAUU ..(((((((((((((((((.(((.......))).))((((....(((((((((....))))))))).))))(..(((((....)))))..)))))))))))))))).............. ( -49.70) >sy_haemo.rRNA 2539906 120 + 2685023 UUGAAAUUCGGCACAGCUUGUACAGGAUAGGUAGGAGCCUUUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACU ..((((..((((((((((.(.............((.(((((....((((((((....)))))))).))))).))(((((....)))))).))))))))))..)))).............. ( -45.50) >sy_aureus.rRNA 2276124 120 + 2809430 UUGAAAUUCGGCACAGCUUGUACAGGAUAGGUAGGAGCCUUUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACU ..((((..((((((((((.(.............((.(((((....((((((((....)))))))).))))).))(((((....)))))).))))))))))..)))).............. ( -45.50) >consensus UUGAAAUUCGGCACAGCUUGUACAGGAUAGGUAGGAGCCUUUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACU ..((((.((((((((((((.(((.......))).))(((((....((((((((....)))))))).)))))...(((((....)))))...)))))))))).)))).............. (-43.63 = -43.97 + 0.33) # Strand winner: reverse (1.00)


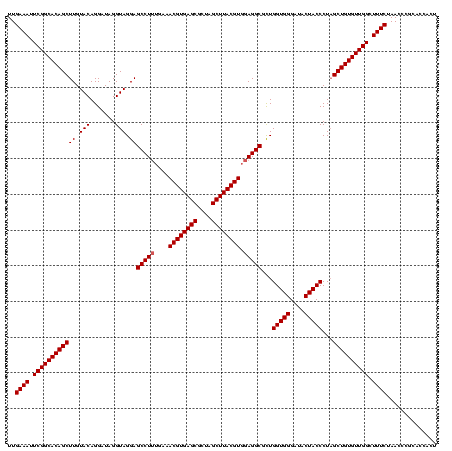
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Fri Apr 7 15:52:15 2006