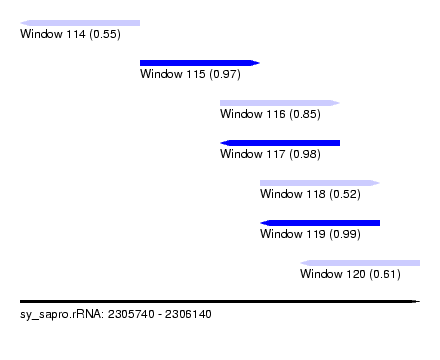
| Sequence ID | sy_sapro.rRNA |
|---|---|
| Location | 2,305,740 – 2,306,140 |
| Length | 400 |
| Max. P | 0.994136 |

| Location | 2,305,740 – 2,305,860 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -38.97 |
| Consensus MFE | -39.63 |
| Energy contribution | -38.97 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.30 |
| Structure conservation index | 1.02 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2305740 120 - 2516583 CUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGGCGAAAGCCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGGUUUCGGCUCGUAAAACUCUGUUAUUAGGGAAGAACAAACGUGU ((((...........))))....((((((.....((((....))))(((((((...((.(((....))).))..((((....))))......))))))).....)))))).......... ( -36.70) >sy_haemo.rRNA 2543563 120 + 2685023 CUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGGCGAAAGCCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUCUUCGGAUCGUAAAACUCUGUUAUUAGGGAAGAACAUACGUGU ((((...........))))....((((((.....((((....))))(((((((...((.(((....))).))..((((....))))......))))))).....)))))).......... ( -40.10) >sy_aureus.rRNA 2279811 120 + 2809430 CUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGGCGAAAGCCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUCUUCGGAUCGUAAAACUCUGUUAUUAGGGAAGAACAUAUGUGU ((((...........))))....((((((.....((((....))))(((((((...((.(((....))).))..((((....))))......))))))).....)))))).......... ( -40.10) >consensus CUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGGCGAAAGCCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUCUUCGGAUCGUAAAACUCUGUUAUUAGGGAAGAACAUACGUGU ((((...........))))....((((((.....((((....))))(((((((...((.(((....))).))..((((....))))......))))))).....)))))).......... (-39.63 = -38.97 + -0.66) # Strand winner: reverse (1.00)

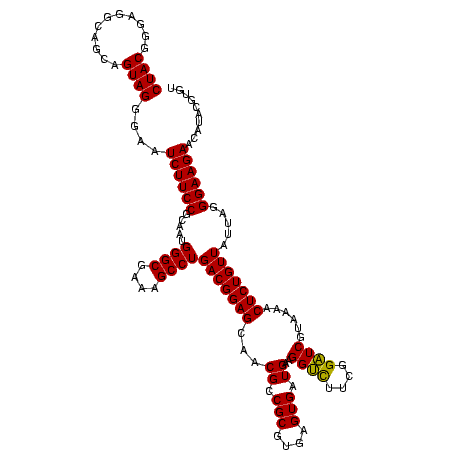
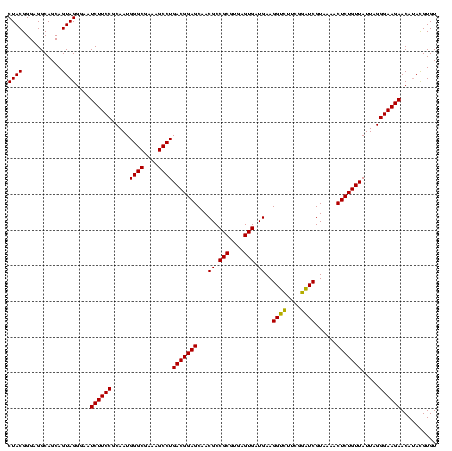
| Location | 2,305,860 – 2,305,980 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -43.21 |
| Consensus MFE | -42.37 |
| Energy contribution | -42.04 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2305860 120 + 2516583 GAGUCUGGACCGUGUCUCAGUUCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGUAUCGUCGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUACGGCGCGGGUCCAUCUAUAA .....(((((((((((...........((((((((((.........))))))))))((((....((.((((((((.......))))))))...))..))))))))).))))))....... ( -43.90) >sy_haemo.rRNA 2543683 120 - 2685023 GAGUCUGGACCGUGUCUCAGUUCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGUAUCGUCGCCUUGGUAAGCCGUUACCUUACCAACUAACUAAUACGGCGCGGGUCCAUCUAUAA .....(((((((((((...........((((((((((.........))))))))))((((.......((((((((.......)))))))).......))))))))).))))))....... ( -43.34) >sy_aureus.rRNA 2279931 120 - 2809430 GAGUCUGGACCGUGUCUCAGUUCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUAUGCAUCGUUGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUGCAGCGCGGAUCCAUCUAUAA .....((((((((((..((......((((((((((((.........))))))))))))...((((..((((((((.......)))))))).))))...))..)))))).))))....... ( -42.40) >consensus GAGUCUGGACCGUGUCUCAGUUCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGUAUCGUCGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUACGGCGCGGGUCCAUCUAUAA .....(((((((((((...........((((((((((.........))))))))))((((.......((((((((.......)))))))).......))))))))))).))))....... (-42.37 = -42.04 + -0.33)

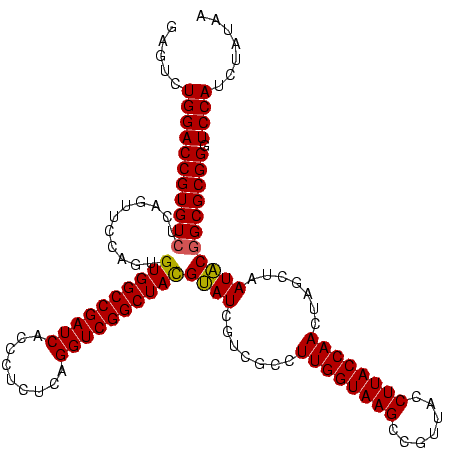
| Location | 2,305,940 – 2,306,060 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -28.23 |
| Energy contribution | -28.23 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849511 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2305940 120 + 2516583 ACCUUACCAACUAGCUAAUACGGCGCGGGUCCAUCUAUAAGUGAUAGCAAAACCAUCUUUCACUUUAGAACCAUGCGGUUCCAAAUGUUAUCCGGUAUUAGCUCCGGUUUCCCGAAGUUA ........(((((((((((((.(.((((((...((((.((((((.((.........)).))))))))))))).)))(((..(....)..)))).))))))))).(((....))).)))). ( -29.70) >sy_haemo.rRNA 2543763 120 - 2685023 ACCUUACCAACUAACUAAUACGGCGCGGGUCCAUCUAUAAGUGAUAGCAAAACCAUCUUUCACUAUCGAACCAUGCGGUUCGAAAUAUUAUCCGGUAUUAGCUCCGGUUUCCCGAAGUUA ........((((..(((((((....(((((.........(((((.((.........)).))))).(((((((....)))))))......))))))))))))...(((....))).)))). ( -28.20) >sy_aureus.rRNA 2280011 120 - 2809430 ACCUUACCAACUAGCUAAUGCAGCGCGGAUCCAUCUAUAAGUGACAGCAAGACCGUCUUUCACUUUUGAACCAUGCGGUUCAAAAUAUUAUCCGGUAUUAGCUCCGGUUUCCCGAAGUUA ........(((((((((((((....(((((..........((((.(((......).)).))))(((((((((....)))))))))....)))))))))))))).(((....))).)))). ( -33.00) >consensus ACCUUACCAACUAGCUAAUACGGCGCGGGUCCAUCUAUAAGUGAUAGCAAAACCAUCUUUCACUUUAGAACCAUGCGGUUCAAAAUAUUAUCCGGUAUUAGCUCCGGUUUCCCGAAGUUA ........(((((((((((((....(((((.........(((((.((.........)).))))).(((((((....)))))))......)))))))))))))).(((....))).)))). (-28.23 = -28.23 + 0.01)

| Location | 2,305,940 – 2,306,060 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -38.50 |
| Consensus MFE | -37.52 |
| Energy contribution | -36.63 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2305940 120 - 2516583 UAACUUCGGGAAACCGGAGCUAAUACCGGAUAACAUUUGGAACCGCAUGGUUCUAAAGUGAAAGAUGGUUUUGCUAUCACUUAUAGAUGGACCCGCGCCGUAUUAGCUAGUUGGUAAGGU (((((((((....))))(((((((((.((......(((((((((....)))))))))(((...(((((.....))))).((.......))...))).))))))))))))))))....... ( -38.90) >sy_haemo.rRNA 2543763 120 + 2685023 UAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUCGAACCGCAUGGUUCGAUAGUGAAAGAUGGUUUUGCUAUCACUUAUAGAUGGACCCGCGCCGUAUUAGUUAGUUGGUAAGGU (((((((((....))))(((((((((((((......))))...(((..((((((((((..(((.....)))..))))).(.....)..))))).)))..))))))))))))))....... ( -37.20) >sy_aureus.rRNA 2280011 120 + 2809430 UAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUUGAACCGCAUGGUUCAAAAGUGAAAGACGGUCUUGCUGUCACUUAUAGAUGGAUCCGCGCUGCAUUAGCUAGUUGGUAAGGU ...((((((....))))))....((((((((....(((((((((....)))))))))((((..(((((.....)))))..))))......))))..(((((....)).))).)))).... ( -39.40) >consensus UAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUCGAACCGCAUGGUUCAAAAGUGAAAGAUGGUUUUGCUAUCACUUAUAGAUGGACCCGCGCCGUAUUAGCUAGUUGGUAAGGU (((((((((....))))(((((((((.((......(((((((((....)))))))))(((...(((((.....))))).((.......))...))).))))))))))))))))....... (-37.52 = -36.63 + -0.88) # Strand winner: reverse (1.00)

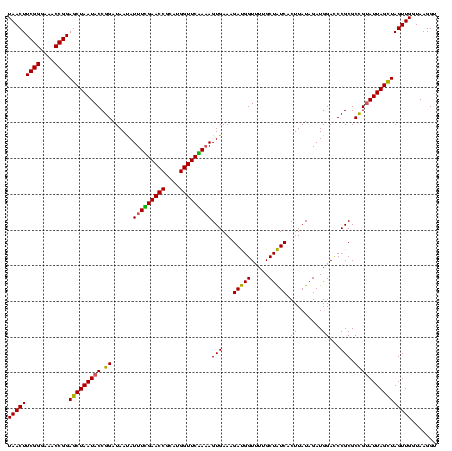
| Location | 2,305,980 – 2,306,100 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -29.53 |
| Consensus MFE | -27.92 |
| Energy contribution | -28.70 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.95 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2305980 120 + 2516583 GUGAUAGCAAAACCAUCUUUCACUUUAGAACCAUGCGGUUCCAAAUGUUAUCCGGUAUUAGCUCCGGUUUCCCGAAGUUAUCCCAGUCUUAUAGGUAGGUUACCCACGUGUUACUCACCC (((((((((..(((.......(((((.(((((....))))).))).)).....((...(((((.(((....))).)))))..)).........))).((....))...))))).)))).. ( -26.40) >sy_haemo.rRNA 2543803 120 - 2685023 GUGAUAGCAAAACCAUCUUUCACUAUCGAACCAUGCGGUUCGAAAUAUUAUCCGGUAUUAGCUCCGGUUUCCCGAAGUUAUCCCAGUCUUAUAGGUAGGUUACCCACGUGUUACUCACCC (((((((((.((((((((...(((.(((((((....)))))))..........((...(((((.(((....))).)))))..))))).....)))).)))).......))))).)))).. ( -31.20) >sy_aureus.rRNA 2280051 120 - 2809430 GUGACAGCAAGACCGUCUUUCACUUUUGAACCAUGCGGUUCAAAAUAUUAUCCGGUAUUAGCUCCGGUUUCCCGAAGUUAUCCCAGUCUUAUAGGUAGGUUAUCCACGUGUUACUCACCC ((((((..(((((..........(((((((((....)))))))))........((...(((((.(((....))).)))))..)).)))))....((.((....)))).))))))...... ( -31.00) >consensus GUGAUAGCAAAACCAUCUUUCACUUUAGAACCAUGCGGUUCAAAAUAUUAUCCGGUAUUAGCUCCGGUUUCCCGAAGUUAUCCCAGUCUUAUAGGUAGGUUACCCACGUGUUACUCACCC (((((((((..(((.........(((((((((....)))))))))........((...(((((.(((....))).)))))..)).........))).((....))...))))).)))).. (-27.92 = -28.70 + 0.78)

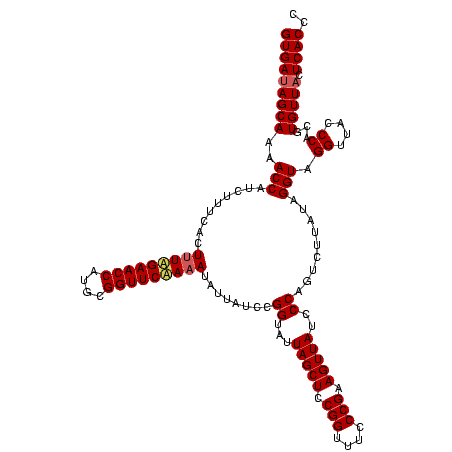
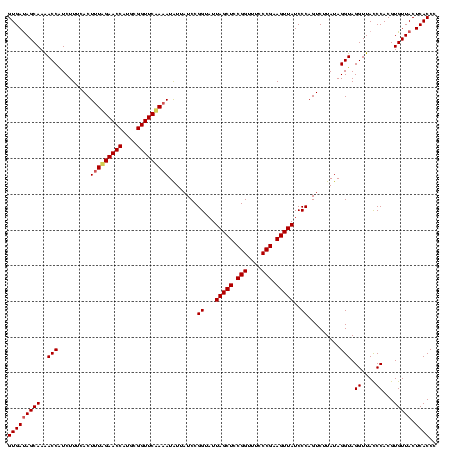
| Location | 2,305,980 – 2,306,100 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -39.63 |
| Consensus MFE | -39.44 |
| Energy contribution | -39.00 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.98 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2305980 120 - 2516583 GGGUGAGUAACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAACAUUUGGAACCGCAUGGUUCUAAAGUGAAAGAUGGUUUUGCUAUCAC ..........((((((((((...)))))))....((((..((.((((((....)))))).))...))))......(((((((((....))))))))))))...(((((.....))))).. ( -40.40) >sy_haemo.rRNA 2543803 120 + 2685023 GGGUGAGUAACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUCGAACCGCAUGGUUCGAUAGUGAAAGAUGGUUUUGCUAUCAC ..........((((((((((...)))))))....((((..((.((((((....)))))).))...))))........(((((((....)))))))..)))...(((((.....))))).. ( -40.00) >sy_aureus.rRNA 2280051 120 + 2809430 GGGUGAGUAACACGUGGAUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUUGAACCGCAUGGUUCAAAAGUGAAAGACGGUCUUGCUGUCAC ..........(((((((.....))))........((((..((.((((((....)))))).))...))))......(((((((((....))))))))))))...(((((.....))))).. ( -38.50) >consensus GGGUGAGUAACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUCGAACCGCAUGGUUCAAAAGUGAAAGAUGGUUUUGCUAUCAC ..........((((((((((...)))))))....((((..((.((((((....)))))).))...))))......(((((((((....))))))))))))...(((((.....))))).. (-39.44 = -39.00 + -0.44) # Strand winner: reverse (1.00)

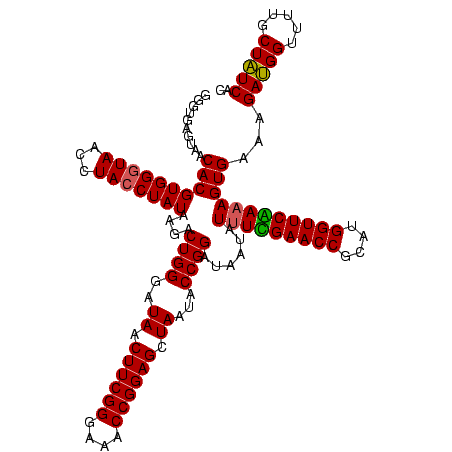
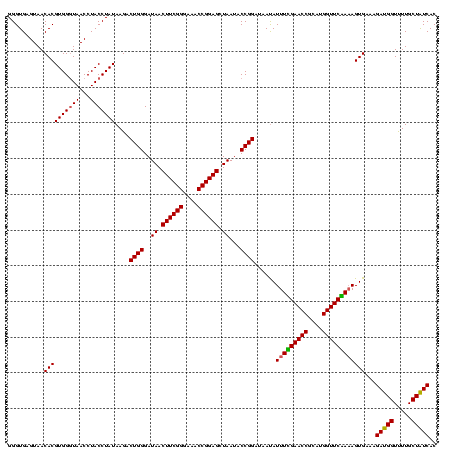
| Location | 2,306,020 – 2,306,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -37.93 |
| Consensus MFE | -34.93 |
| Energy contribution | -34.60 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2306020 120 - 2516583 AACAGAUAAGGAGCUUGCUCCUUUGACGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAACAUUUGG ..(((((..(..(((..(((.((((.((....)).)))).)))..)))..)..(((((((...)))))))....((((..((.((((((....)))))).))...)))).....))))). ( -39.70) >sy_haemo.rRNA 2543843 120 + 2685023 AACAGACAAGGAGCUUGCUCCUUUGACGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUCG .........(..(((..(((.((((.((....)).)))).)))..)))..)..(((((((...)))))))....((((..((.((((((....)))))).))...))))........... ( -37.30) >sy_aureus.rRNA 2280091 120 + 2809430 AACGGACGAGAAGCUUGCUUCUCUGAUGUUAGCGGCGGACGGGUGAGUAACACGUGGAUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUUG ..(((..((((((....))))))...(((((((((....((.(((.....))).)).....))..((((......)))).....(((((....)))))))))))))))............ ( -36.80) >consensus AACAGACAAGGAGCUUGCUCCUUUGACGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUCG .........(..(((..(((.((((.((....)).)))).)))..)))..)..(((((((...)))))))....((((..((.((((((....)))))).))...))))........... (-34.93 = -34.60 + -0.33) # Strand winner: reverse (1.00)

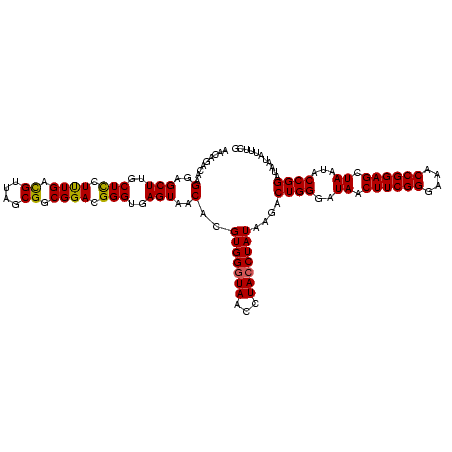
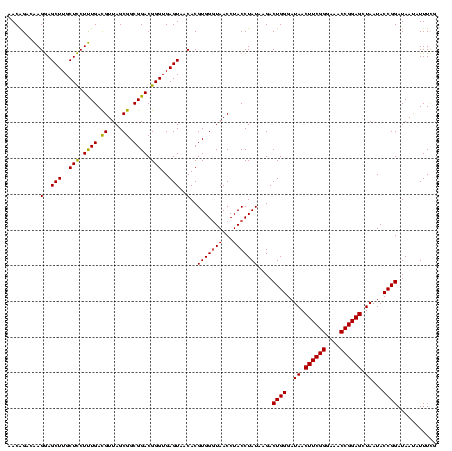
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Fri Apr 7 15:52:07 2006