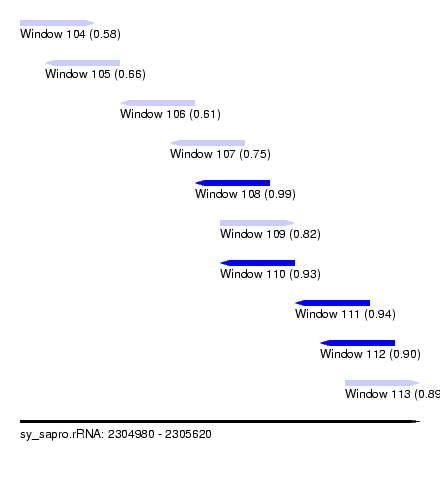
| Sequence ID | sy_sapro.rRNA |
|---|---|
| Location | 2,304,980 – 2,305,620 |
| Length | 640 |
| Max. P | 0.989071 |

| Location | 2,304,980 – 2,305,100 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -36.50 |
| Energy contribution | -36.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.39 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.579190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2304980 120 + 2516583 AAAUCAUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCAACCUAGAGUGCCCAACUUAAUGAUGGCAACUAAGCUUAAGGGUUGCGCUCGUUGC .........((((.((((((......))))))))))......(((((((.....))))).)).((((...((((((....(((((.(..(....)....))))))....)))))))))). ( -36.50) >sy_haemo.rRNA 2542803 120 - 2685023 AAAUCAUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCAACUUAGAGUGCCCAACUUAAUGAUGGCAACUAAGCUUAAGGGUUGCGCUCGUUGC .........((((.((((((......))))))))))......(((((((.....))))).)).((((...((((((....(((((.(..(....)....))))))....)))))))))). ( -36.50) >sy_aureus.rRNA 2279051 120 - 2809430 AAAUCAUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCAACUUAGAGUGCCCAACUUAAUGAUGGCAACUAAGCUUAAGGGUUGCGCUCGUUGC .........((((.((((((......))))))))))......(((((((.....))))).)).((((...((((((....(((((.(..(....)....))))))....)))))))))). ( -36.50) >consensus AAAUCAUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCAACUUAGAGUGCCCAACUUAAUGAUGGCAACUAAGCUUAAGGGUUGCGCUCGUUGC .........((((.((((((......))))))))))......(((((((.....))))).)).((((...((((((....(((((.(..(....)....))))))....)))))))))). (-36.50 = -36.50 + 0.00)

| Location | 2,305,020 – 2,305,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -42.10 |
| Consensus MFE | -42.32 |
| Energy contribution | -42.10 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.48 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2305020 120 - 2516583 UUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAGGUUGACUGCCGGUGACAAACCGGAGGA .....(((.(((((((((....)))((.(((......))))).))))))..........(((((((.((((..(.....)...))))..))))))))))..(((((.....))))).... ( -41.90) >sy_haemo.rRNA 2542843 120 + 2685023 UUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAAGUUGACUGCCGGUGACAAACCGGAGGA .....(((.(((((((((....)))((.(((......))))).))))))..........(((((((.((((..(.....)...))))..))))))))))..(((((.....))))).... ( -42.20) >sy_aureus.rRNA 2279091 120 + 2809430 UUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAAGUUGACUGCCGGUGACAAACCGGAGGA .....(((.(((((((((....)))((.(((......))))).))))))..........(((((((.((((..(.....)...))))..))))))))))..(((((.....))))).... ( -42.20) >consensus UUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAAGUUGACUGCCGGUGACAAACCGGAGGA .....(((.(((((((((....)))((.(((......))))).))))))..........(((((((.((((..(.....)...))))..))))))))))..(((((.....))))).... (-42.32 = -42.10 + -0.22) # Strand winner: reverse (0.99)

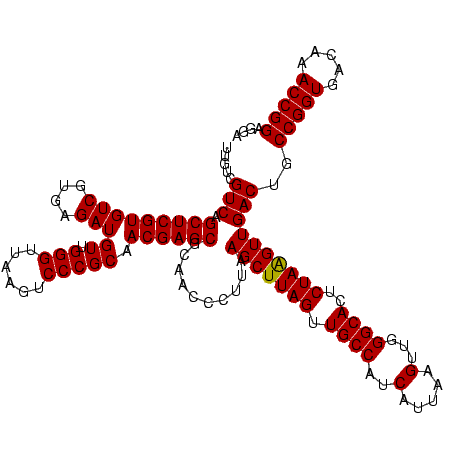
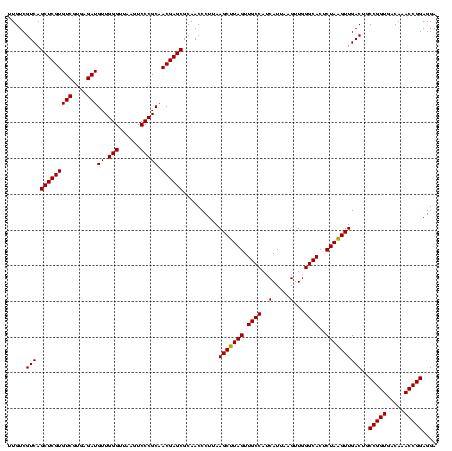
| Location | 2,305,140 – 2,305,260 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -37.90 |
| Consensus MFE | -37.90 |
| Energy contribution | -37.90 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.44 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.614695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2305140 120 - 2516583 GGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUUACCAAAUCUUGACAUCCUUUGAAAACUCUAGAGAUAGAGCCUUCCCCUUCGGGGGACAAAGUGACAGGUGGUGCAUGG .......((((((((((..((((........)))))))))..((((...((((...(((((((....(((((....)))))...(((((....)))))))))).))))))))))))))). ( -37.90) >sy_haemo.rRNA 2542963 120 + 2685023 GGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUUACCAAAUCUUGACAUCCUUUGACAACUCUAGAGAUAGAGCCUUCCCCUUCGGGGGACAAAGUGACAGGUGGUGCAUGG .......((((((((((..((((........)))))))))..((((...((((...(((((((....(((((....)))))...(((((....)))))))))).))))))))))))))). ( -37.90) >sy_aureus.rRNA 2279211 120 + 2809430 GGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUUACCAAAUCUUGACAUCCUUUGACAACUCUAGAGAUAGAGCCUUCCCCUUCGGGGGACAAAGUGACAGGUGGUGCAUGG .......((((((((((..((((........)))))))))..((((...((((...(((((((....(((((....)))))...(((((....)))))))))).))))))))))))))). ( -37.90) >consensus GGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUUACCAAAUCUUGACAUCCUUUGACAACUCUAGAGAUAGAGCCUUCCCCUUCGGGGGACAAAGUGACAGGUGGUGCAUGG .......((((((((((..((((........)))))))))..((((...((((...(((((((....(((((....)))))...(((((....)))))))))).))))))))))))))). (-37.90 = -37.90 + -0.00) # Strand winner: reverse (0.98)

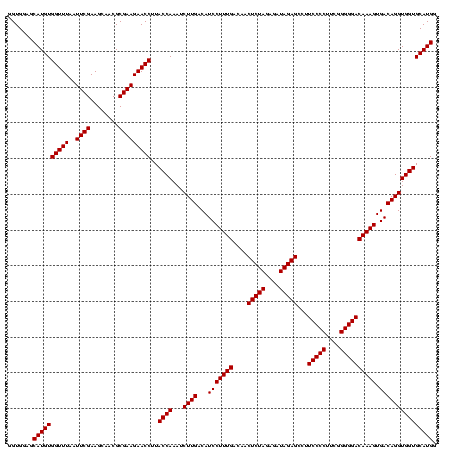
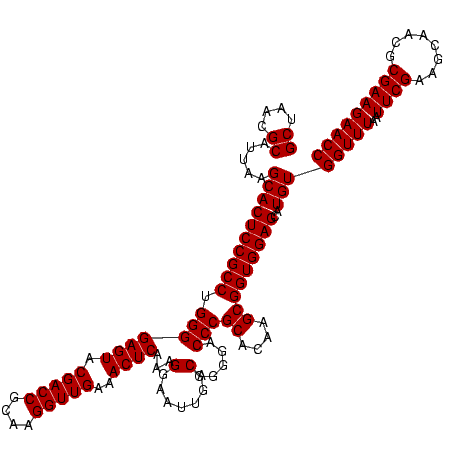
| Location | 2,305,220 – 2,305,340 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -40.10 |
| Consensus MFE | -40.10 |
| Energy contribution | -40.10 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.746171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2305220 120 - 2516583 GCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACC ((....)).....((((((((((.(((((((.(((((....)))))..))))...(.......).....)))((....)))))))))..)))(((((..((((........))))))))) ( -40.10) >sy_haemo.rRNA 2543043 120 + 2685023 GCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACC ((....)).....((((((((((.(((((((.(((((....)))))..))))...(.......).....)))((....)))))))))..)))(((((..((((........))))))))) ( -40.10) >sy_aureus.rRNA 2279291 120 + 2809430 GCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACC ((....)).....((((((((((.(((((((.(((((....)))))..))))...(.......).....)))((....)))))))))..)))(((((..((((........))))))))) ( -40.10) >consensus GCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACC ((....)).....((((((((((.(((((((.(((((....)))))..))))...(.......).....)))((....)))))))))..)))(((((..((((........))))))))) (-40.10 = -40.10 + 0.00) # Strand winner: reverse (0.86)

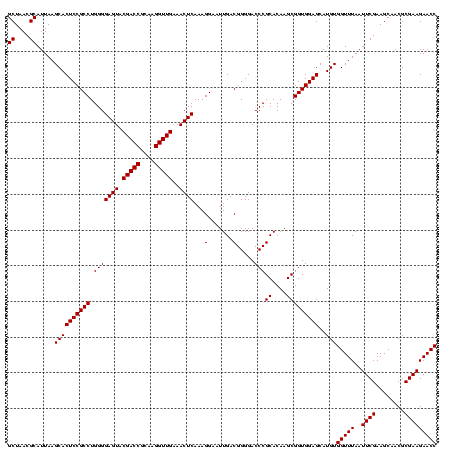
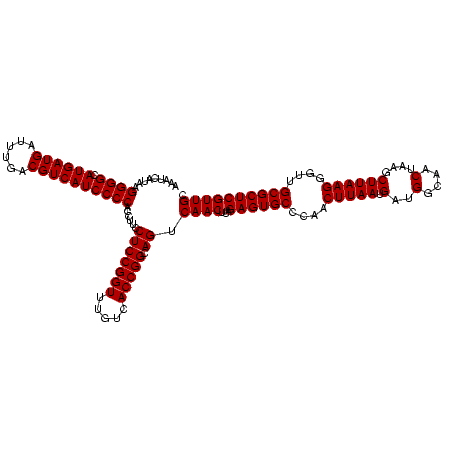
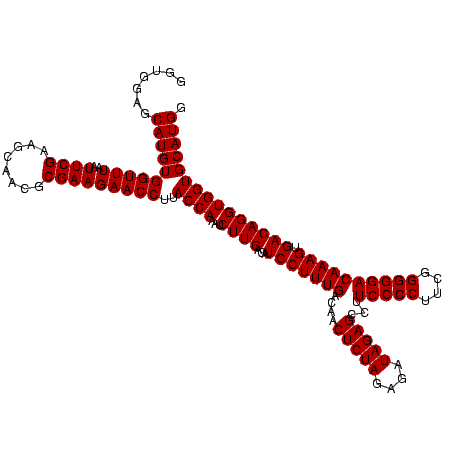
| Location | 2,305,260 – 2,305,380 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -50.20 |
| Consensus MFE | -50.20 |
| Energy contribution | -50.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.16 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2305260 120 - 2516583 AGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGC ((((((.((..(((((((((....)))))))))..))...((....))....))))))...(((...((((.(((((....)))))..))))..)))...((.(((....)))))..... ( -50.20) >sy_haemo.rRNA 2543083 120 + 2685023 AGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGC ((((((.((..(((((((((....)))))))))..))...((....))....))))))...(((...((((.(((((....)))))..))))..)))...((.(((....)))))..... ( -50.20) >sy_aureus.rRNA 2279331 120 + 2809430 AGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGC ((((((.((..(((((((((....)))))))))..))...((....))....))))))...(((...((((.(((((....)))))..))))..)))...((.(((....)))))..... ( -50.20) >consensus AGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGC ((((((.((..(((((((((....)))))))))..))...((....))....))))))...(((...((((.(((((....)))))..))))..)))...((.(((....)))))..... (-50.20 = -50.20 + -0.00) # Strand winner: reverse (0.92)

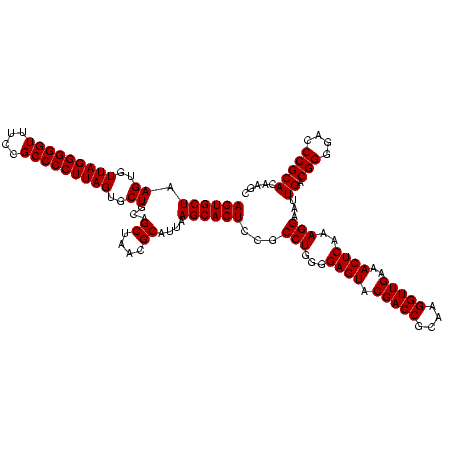
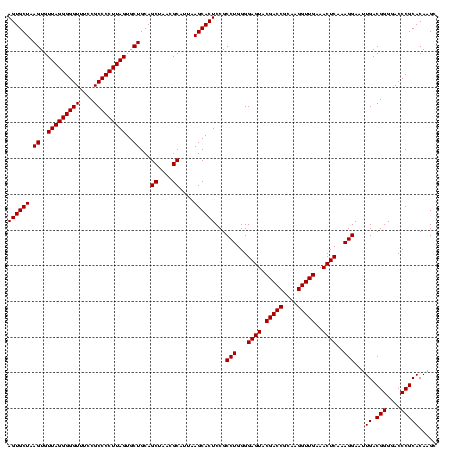
| Location | 2,305,300 – 2,305,420 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -43.80 |
| Consensus MFE | -43.80 |
| Energy contribution | -43.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2305300 120 + 2516583 UGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGU ...((((.(((.((..((((((((((((....(.((((((....)).....((((.(....))))))))).)..)))))))..)))))..)).)))))))..((((((.....)))))). ( -43.80) >sy_haemo.rRNA 2543123 120 - 2685023 UGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGU ...((((.(((.((..((((((((((((....(.((((((....)).....((((.(....))))))))).)..)))))))..)))))..)).)))))))..((((((.....)))))). ( -43.80) >sy_aureus.rRNA 2279371 120 - 2809430 UGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGU ...((((.(((.((..((((((((((((....(.((((((....)).....((((.(....))))))))).)..)))))))..)))))..)).)))))))..((((((.....)))))). ( -43.80) >consensus UGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGU ...((((.(((.((..((((((((((((....(.((((((....)).....((((.(....))))))))).)..)))))))..)))))..)).)))))))..((((((.....)))))). (-43.80 = -43.80 + 0.00)

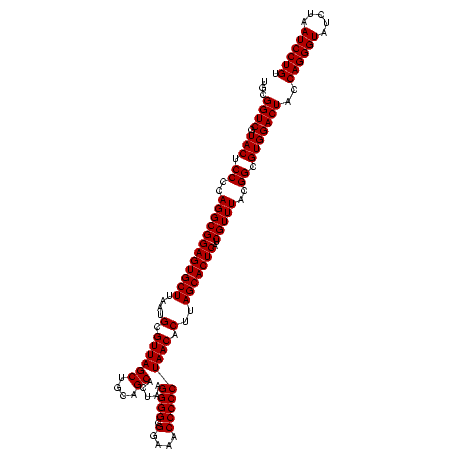
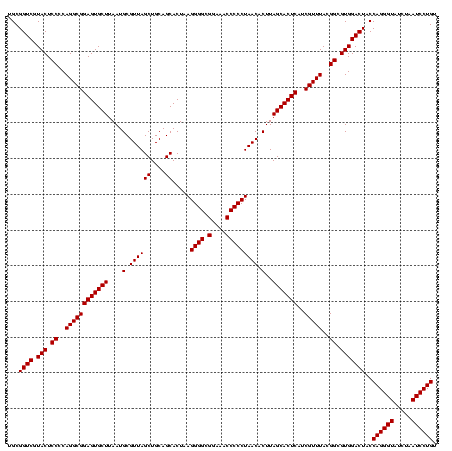
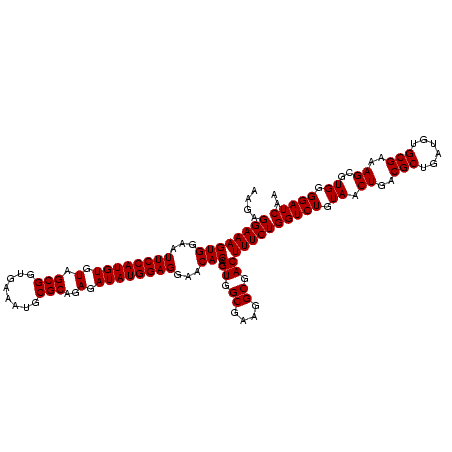
| Location | 2,305,300 – 2,305,420 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -46.00 |
| Consensus MFE | -46.00 |
| Energy contribution | -46.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.934425 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2305300 120 - 2516583 ACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCA .((((.........))))(..(((.((.((.((..((..(((((((.((..(((((((((....)))))))))..))...((....))....)))))))))..)).)).))..)))..). ( -46.00) >sy_haemo.rRNA 2543123 120 + 2685023 ACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCA .((((.........))))(..(((.((.((.((..((..(((((((.((..(((((((((....)))))))))..))...((....))....)))))))))..)).)).))..)))..). ( -46.00) >sy_aureus.rRNA 2279371 120 + 2809430 ACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCA .((((.........))))(..(((.((.((.((..((..(((((((.((..(((((((((....)))))))))..))...((....))....)))))))))..)).)).))..)))..). ( -46.00) >consensus ACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCA .((((.........))))(..(((.((.((.((..((..(((((((.((..(((((((((....)))))))))..))...((....))....)))))))))..)).)).))..)))..). (-46.00 = -46.00 + 0.00) # Strand winner: reverse (0.81)



| Location | 2,305,420 – 2,305,540 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -36.90 |
| Consensus MFE | -36.90 |
| Energy contribution | -36.90 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.17 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2305420 120 - 2516583 AAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACGCUGAUGUGCGAAAGCGUGGGGAUCAA ....((((((((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).)))))))(((((.((.((..(((......)))..))..)).))))).. ( -36.90) >sy_haemo.rRNA 2543243 120 + 2685023 AAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACGCUGAUGUGCGAAAGCGUGGGGAUCAA ....((((((((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).)))))))(((((.((.((..(((......)))..))..)).))))).. ( -36.90) >sy_aureus.rRNA 2279491 120 + 2809430 AAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACGCUGAUGUGCGAAAGCGUGGGGAUCAA ....((((((((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).)))))))(((((.((.((..(((......)))..))..)).))))).. ( -36.90) >consensus AAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACGCUGAUGUGCGAAAGCGUGGGGAUCAA ....((((((((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).)))))))(((((.((.((..(((......)))..))..)).))))).. (-36.90 = -36.90 + -0.00) # Strand winner: reverse (1.00)

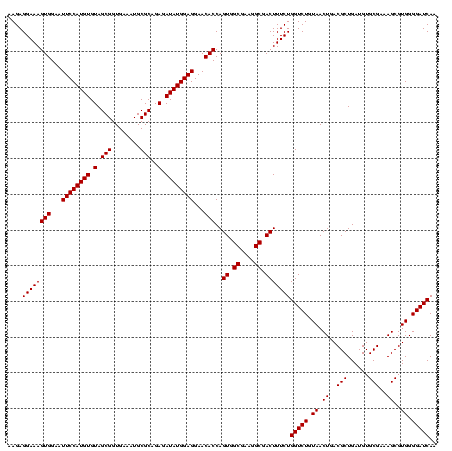
| Location | 2,305,460 – 2,305,580 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -34.07 |
| Energy contribution | -34.07 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2305460 120 - 2516583 ACCGUGGAGGGUCAUUGGAAACUGGGAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUG .....((((((((.......(((((....)...................(((...((((((((.(.(((........)))..).))))))))...))))))).((....)))))))))). ( -34.90) >sy_haemo.rRNA 2543283 120 + 2685023 ACCGUGGAGGGUCAUUGGAAACUGGAAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUG .....((((((((.......(((((....(((........)))......((....((((((((.(.(((........)))..).))))))))..)).))))).((....)))))))))). ( -34.00) >sy_aureus.rRNA 2279531 120 + 2809430 ACCGUGGAGGGUCAUUGGAAACUGGAAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUG .....((((((((.......(((((....(((........)))......((....((((((((.(.(((........)))..).))))))))..)).))))).((....)))))))))). ( -34.00) >consensus ACCGUGGAGGGUCAUUGGAAACUGGAAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUG .....((((((((.......(((((....(((........)))......((....((((((((.(.(((........)))..).))))))))..)).))))).((....)))))))))). (-34.07 = -34.07 + 0.00) # Strand winner: reverse (1.00)

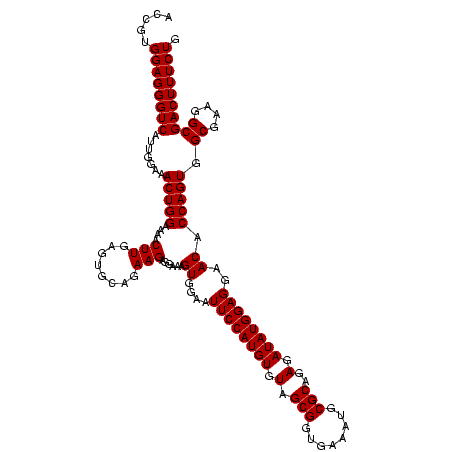
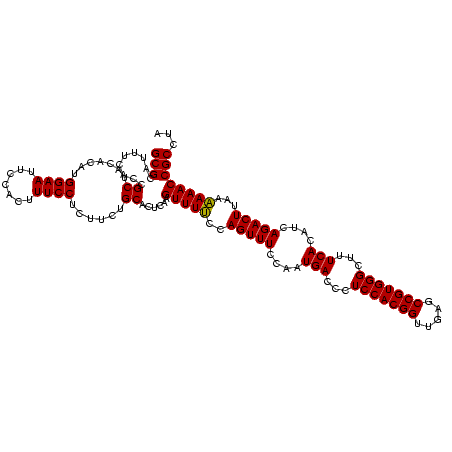
| Location | 2,305,500 – 2,305,620 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -25.95 |
| Consensus MFE | -26.40 |
| Energy contribution | -25.95 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.85 |
| Structure conservation index | 1.02 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 2305500 120 + 2516583 GCGCAUUUCACCGCUACACAUGGAAUUCCACUUUCCUCUUCUGCACUCAAGUUUCCCAGUUUCCAAUGACCCUCCACGGUUGAGCCGUGGGCUUUCACAUCAGACUUAAGAAACCGCCUA (((.........((.......((((.......))))......))......(((((..(((((....(((...(((((((.....)))))))...)))....)))))...))))))))... ( -27.42) >sy_haemo.rRNA 2543323 120 - 2685023 GCGCAUUUCACCGCUACACAUGGAAUUCCACUUUCCUCUUCUGCACUCAAGUUUUCCAGUUUCCAAUGACCCUCCACGGUUGAGCCGUGGGCUUUCACAUCAGACUUAAAAAACCGCCUA (((.........((.......((((.......))))......))......(((((..(((((....(((...(((((((.....)))))))...)))....)))))...))))))))... ( -25.22) >sy_aureus.rRNA 2279571 120 - 2809430 GCGCAUUUCACCGCUACACAUGGAAUUCCACUUUCCUCUUCUGCACUCAAGUUUUCCAGUUUCCAAUGACCCUCCACGGUUGAGCCGUGGGCUUUCACAUCAGACUUAAAAAACCGCCUA (((.........((.......((((.......))))......))......(((((..(((((....(((...(((((((.....)))))))...)))....)))))...))))))))... ( -25.22) >consensus GCGCAUUUCACCGCUACACAUGGAAUUCCACUUUCCUCUUCUGCACUCAAGUUUUCCAGUUUCCAAUGACCCUCCACGGUUGAGCCGUGGGCUUUCACAUCAGACUUAAAAAACCGCCUA (((.........((.......((((.......))))......))......(((((..(((((....(((...(((((((.....)))))))...)))....)))))...))))))))... (-26.40 = -25.95 + -0.44)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Fri Apr 7 15:52:01 2006