
| Sequence ID | sy_aureus.rRNA |
|---|---|
| Location | 579,928 – 580,286 |
| Length | 358 |
| Max. P | 0.999811 |

| Location | 579,928 – 580,048 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.40 |
| Mean single sequence MFE | -35.53 |
| Consensus MFE | -26.04 |
| Energy contribution | -25.35 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.73 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
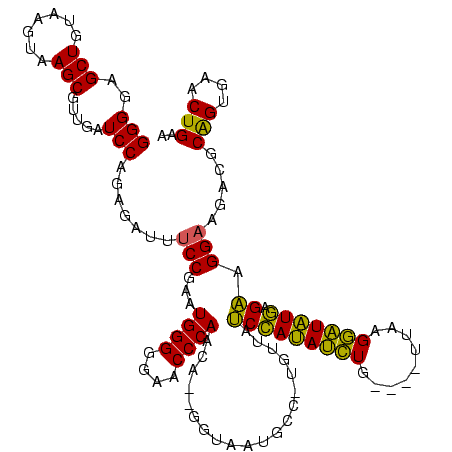
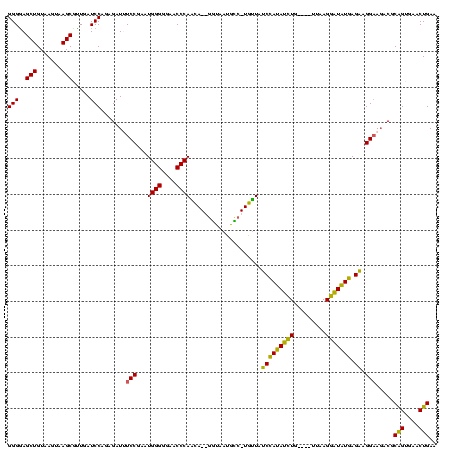
>sy_aureus.rRNA 579928 120 + 2809430 GGGGAGCUGUAAGUAAGCUUUGAUCCAGAGAUUUCCGAAUGGGGAAACCCAGCAUGAGUUAUGUCAUGUUAUCGAUAUGUGAAUACAUAGCAUAUCAGAAGGCACACCCGGAGAACUGAA ((((((((.......)))))...)))..((.((((((...(((....)))(((((((......)))))))...(((((((.........)))))))............)))))).))... ( -35.80) >sp_agal.rRNA 18317 113 + 2160275 GGGGAGCUGUAAGUAAGCGCUGAUCCAGAGAUGUCCGAAUGGGGGAACCCACUA--GUUAAUGAC-UAGUAUCCAUAUCUG----UUAAGGAUAUGAGAAGGAAGACGCAGUGAACUGAA (((.(((.((......)))))..))).......(((....(((....)))((((--(((...)))-)))).(((((((((.----....))))))).)).))).....(((....))).. ( -32.20) >sp_mutans.rRNA 18898 113 + 2030929 GGGGAGCUGUAAGUAAGCCUUGAUCCAGAGAUAUCCGAAUGGGGGAACCCAACA--GGUAAUGCC-UGUUAUCCAUAACUG----UUAAGGUUAUGAGAAGGAAGACGCAGUGAACUGAA .((..(((((..((...((((........((((......((((....))))(((--((.....))-)))))))(((((((.----....)))))))..))))...)))))))...))... ( -33.40) >sp_pneumo.rRNA 17028 113 + 2038623 GGGUAGCUGUAAGUAAGCGAUGAUCCAGGGAUUUCCGAAUGGGGGAACCCAACA--GGUAAUACC-UGUUACCCACAUCUG----UUAAGGAUGUGAGGAGGAAGACGCAGUGAACUGAA ((((.(((.......)))....)))).....(((((....(((....)))((((--((.....))-)))).(((((((((.----....))))))).)).)))))...(((....))).. ( -40.70) >consensus GGGGAGCUGUAAGUAAGCGUUGAUCCAGAGAUUUCCGAAUGGGGGAACCCAACA__GGUAAUGCC_UGUUAUCCAUAUCUG____UUAAGGAUAUGAGAAGGAAGACGCAGUGAACUGAA (((..(((.......))).....))).......(((...((((....))))....................(((((((((.........))))))).)).))).....(((....))).. (-26.04 = -25.35 + -0.69)

| Location | 579,928 – 580,048 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.40 |
| Mean single sequence MFE | -25.68 |
| Consensus MFE | -16.94 |
| Energy contribution | -14.88 |
| Covariance contribution | -2.06 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 579928 120 - 2809430 UUCAGUUCUCCGGGUGUGCCUUCUGAUAUGCUAUGUAUUCACAUAUCGAUAACAUGACAUAACUCAUGCUGGGUUUCCCCAUUCGGAAAUCUCUGGAUCAAAGCUUACUUACAGCUCCCC (((((...((((((((.((((..(((((((...........)))))))....(((((......)))))..)))).....)))))))).....)))))....((((.......)))).... ( -25.90) >sp_agal.rRNA 18317 113 - 2160275 UUCAGUUCACUGCGUCUUCCUUCUCAUAUCCUUAA----CAGAUAUGGAUACUA-GUCAUUAAC--UAGUGGGUUCCCCCAUUCGGACAUCUCUGGAUCAGCGCUUACUUACAGCUCCCC ....((.....((((..(((....((((((.....----..))))))(((((((-((.....))--))))(((....)))........)))...)))...))))......))........ ( -23.00) >sp_mutans.rRNA 18898 113 - 2030929 UUCAGUUCACUGCGUCUUCCUUCUCAUAACCUUAA----CAGUUAUGGAUAACA-GGCAUUACC--UGUUGGGUUCCCCCAUUCGGAUAUCUCUGGAUCAAGGCUUACUUACAGCUCCCC ..(((....))).(((((...((.((((((.....----..)))))))).((((-((.....))--))))(((....)))(((((((....))))))).)))))................ ( -23.50) >sp_pneumo.rRNA 17028 113 - 2038623 UUCAGUUCACUGCGUCUUCCUCCUCACAUCCUUAA----CAGAUGUGGGUAACA-GGUAUUACC--UGUUGGGUUCCCCCAUUCGGAAAUCCCUGGAUCAUCGCUUACUUACAGCUACCC ....((.....(((...(((..((((((((.....----..)))))))).((((-((.....))--))))(((((((.......))))..))).)))....)))......))........ ( -30.30) >consensus UUCAGUUCACUGCGUCUUCCUUCUCAUAUCCUUAA____CAGAUAUGGAUAACA_GGCAUUACC__UGUUGGGUUCCCCCAUUCGGAAAUCUCUGGAUCAACGCUUACUUACAGCUCCCC (((((....(((.........((.((((((...........))))))))....................((((....))))..)))......))))).....(((.......)))..... (-16.94 = -14.88 + -2.06) # Strand winner: forward (1.00)

| Location | 579,968 – 580,088 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.26 |
| Mean single sequence MFE | -33.88 |
| Consensus MFE | -28.25 |
| Energy contribution | -25.00 |
| Covariance contribution | -3.25 |
| Combinations/Pair | 1.45 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.53 |
| SVM RNA-class probability | 0.999347 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
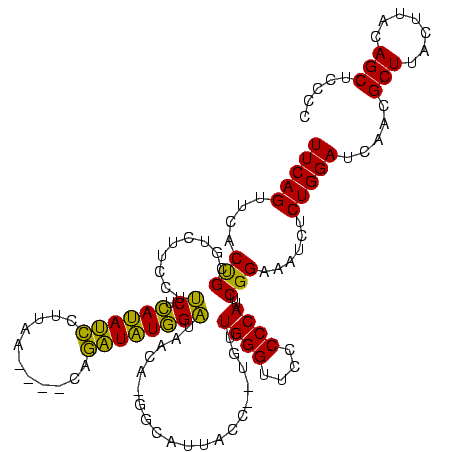
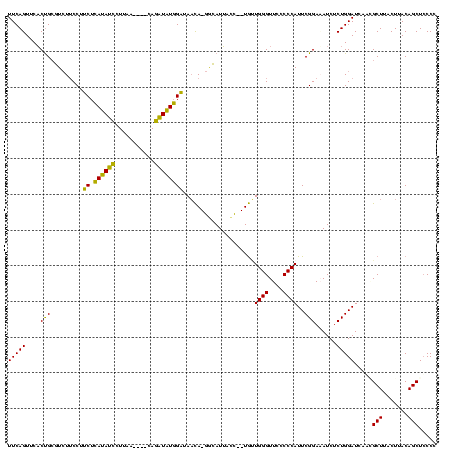
>sy_aureus.rRNA 579968 120 + 2809430 GGGGAAACCCAGCAUGAGUUAUGUCAUGUUAUCGAUAUGUGAAUACAUAGCAUAUCAGAAGGCACACCCGGAGAACUGAAACAUCUUAGUACCCGGAGGAAGAGAAAGAAAAUUCGAUUC (((....)))....((((((...((.(((..(((((((((.........))))))).))..)))(..((((.(.(((((......))))).)))))..)........)).)))))).... ( -33.60) >sp_agal.rRNA 18357 113 + 2160275 GGGGGAACCCACUA--GUUAAUGAC-UAGUAUCCAUAUCUG----UUAAGGAUAUGAGAAGGAAGACGCAGUGAACUGAAACAUCUAAGUAGCUGCAGGAAGAGAAAGCAAAAGCGAUUG (((....)))((((--(((...)))-)))).(((((((((.----....))))))).))........(((((..(((..........))).)))))...........((....))..... ( -29.90) >sp_mutans.rRNA 18938 113 + 2030929 GGGGGAACCCAACA--GGUAAUGCC-UGUUAUCCAUAACUG----UUAAGGUUAUGAGAAGGAAGACGCAGUGAACUGAAACAUCUCAGUAGCUGCAGGAAGAGAAAGCAAGAGCGAUUG (((....)))((((--((.....))-)))).(((((((((.----....))))))).))........(((((..(((((......))))).)))))...........((....))..... ( -36.40) >sp_pneumo.rRNA 17068 113 + 2038623 GGGGGAACCCAACA--GGUAAUACC-UGUUACCCACAUCUG----UUAAGGAUGUGAGGAGGAAGACGCAGUGAACUGAAACAUCUAAGUAGCUGCAGGAAGAGAAAGCAAAAGCGAUUG (((....)))((((--((.....))-)))).(((((((((.----....))))))).))........(((((..(((..........))).)))))...........((....))..... ( -35.60) >consensus GGGGGAACCCAACA__GGUAAUGCC_UGUUAUCCAUAUCUG____UUAAGGAUAUGAGAAGGAAGACGCAGUGAACUGAAACAUCUAAGUAGCUGCAGGAAGAGAAAGCAAAAGCGAUUG (((....)))(((((.((.....))))))).(((((((((.........))))))).))........(((((..(((((......))))).)))))...........((....))..... (-28.25 = -25.00 + -3.25)

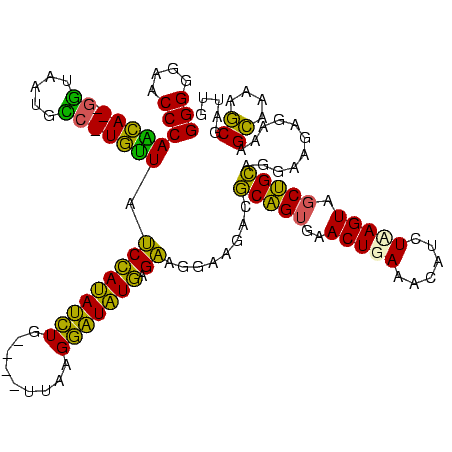
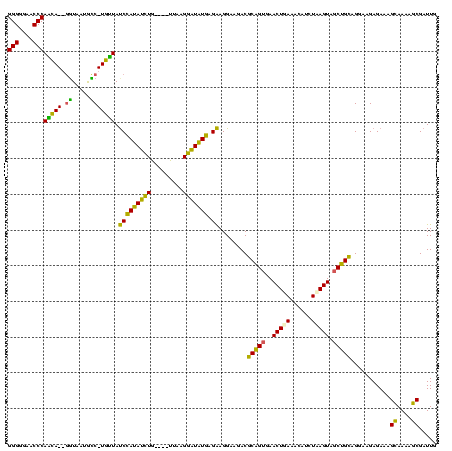
| Location | 580,048 – 580,168 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.15 |
| Mean single sequence MFE | -38.33 |
| Consensus MFE | -31.05 |
| Energy contribution | -27.68 |
| Covariance contribution | -3.37 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 580048 120 + 2809430 ACAUCUUAGUACCCGGAGGAAGAGAAAGAAAAUUCGAUUCCCUUAGUAGCGGCGAGCGAAAUGGGAAGAGCCCAAACCAACAAGCUUGCUUGUUGGGGUUGUAGGACACUCUAUACGGAG ...(((((..((((((.((((..(((......)))..))))))...(((((((((((....((((.....)))).........)))))).)))))))))..)))))..((((....)))) ( -34.12) >sp_agal.rRNA 18430 119 + 2160275 ACAUCUAAGUAGCUGCAGGAAGAGAAAGCAAAAGCGAUUGCCUUAGUAGCGGCGAGCGAAACGGCAGGAGGGCAAACCGAAGUGUUUACACUUCGGGGUUGUAGGAC-UGCGAUAUAGGA ...((((.((.((((((((....((..((....))..)).)))..))))).))..........((((....((((.((((((((....))))))))..))))....)-)))....)))). ( -38.20) >sp_mutans.rRNA 19011 119 + 2030929 ACAUCUCAGUAGCUGCAGGAAGAGAAAGCAAGAGCGAUUGCCUCAGUAGCGGCGAGCGAAGAGGCAGGAGGGCAAACCAGAGUGUUUACACUCUGGGGUUGUAGGAC-UGCGAUAAAGCA ..(((.((((..((((((...............((..(((((((....((.....))...)))))))....))...((((((((....))))))))..)))))).))-)).)))...... ( -42.40) >sp_pneumo.rRNA 17141 119 + 2038623 ACAUCUAAGUAGCUGCAGGAAGAGAAAGCAAAAGCGAUUGCCUUAGUAGCGGCGAGCGAAACGGCAGGAGGGCAAACCGAAGAGUUUACUCUUCGGGGUUGUAGGAC-UGCAAUGUGGAC ...((((.((.((((((((....((..((....))..)).)))..))))).))..........((((....((((.((((((((....))))))))..))))....)-)))....)))). ( -38.60) >consensus ACAUCUAAGUAGCUGCAGGAAGAGAAAGCAAAAGCGAUUGCCUUAGUAGCGGCGAGCGAAACGGCAGGAGGGCAAACCAAAGAGUUUACACUUCGGGGUUGUAGGAC_UGCGAUAUAGAA ...(((...(((((((...........((....))..(((((......((.....)).....)))))....))...((((((((....)))))))))))))..))).............. (-31.05 = -27.68 + -3.37)

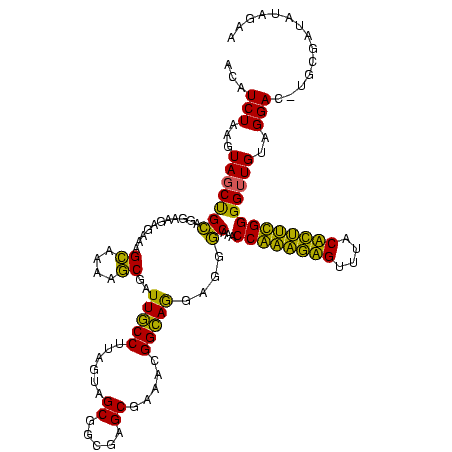
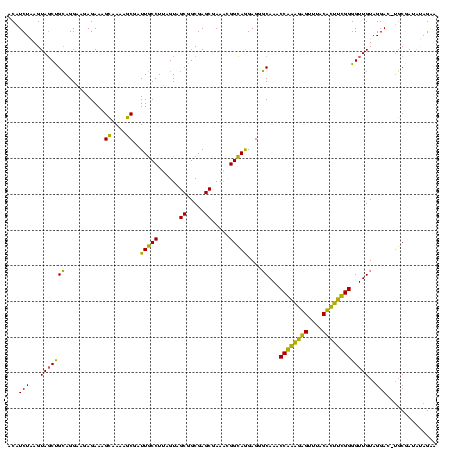
| Location | 580,048 – 580,168 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.15 |
| Mean single sequence MFE | -32.11 |
| Consensus MFE | -25.88 |
| Energy contribution | -22.38 |
| Covariance contribution | -3.50 |
| Combinations/Pair | 1.46 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 580048 120 - 2809430 CUCCGUAUAGAGUGUCCUACAACCCCAACAAGCAAGCUUGUUGGUUUGGGCUCUUCCCAUUUCGCUCGCCGCUACUAAGGGAAUCGAAUUUUCUUUCUCUUCCUCCGGGUACUAAGAUGU ((..((((.(((((..........((((((((....))))))))..((((.....))))...))))).(((.....(((((((..((....)).)))))))....)))))))..)).... ( -32.20) >sp_agal.rRNA 18430 119 - 2160275 UCCUAUAUCGCA-GUCCUACAACCCCGAAGUGUAAACACUUCGGUUUGCCCUCCUGCCGUUUCGCUCGCCGCUACUAAGGCAAUCGCUUUUGCUUUCUCUUCCUGCAGCUACUUAGAUGU ....((((((((-(.....(((..((((((((....)))))))).))).....))))......(((.((.......(((((((......)))))))........)))))......))))) ( -31.06) >sp_mutans.rRNA 19011 119 - 2030929 UGCUUUAUCGCA-GUCCUACAACCCCAGAGUGUAAACACUCUGGUUUGCCCUCCUGCCUCUUCGCUCGCCGCUACUGAGGCAAUCGCUCUUGCUUUCUCUUCCUGCAGCUACUGAGAUGU .....((((.((-((.........((((((((....)))))))).((((.....((((((...((.....))....))))))...((....))...........))))..)))).)))). ( -34.70) >sp_pneumo.rRNA 17141 119 - 2038623 GUCCACAUUGCA-GUCCUACAACCCCGAAGAGUAAACUCUUCGGUUUGCCCUCCUGCCGUUUCGCUCGCCGCUACUAAGGCAAUCGCUUUUGCUUUCUCUUCCUGCAGCUACUUAGAUGU ....((((((((-(.....(((..((((((((....)))))))).))).....))))......(((.((.......(((((((......)))))))........)))))......))))) ( -30.46) >consensus UUCCAUAUCGCA_GUCCUACAACCCCAAAGAGUAAACACUUCGGUUUGCCCUCCUGCCGUUUCGCUCGCCGCUACUAAGGCAAUCGCUUUUGCUUUCUCUUCCUGCAGCUACUUAGAUGU .....((((...............((((((((....)))))))).((((.....((((.....((.....))......))))...((....))...........)))).......)))). (-25.88 = -22.38 + -3.50) # Strand winner: forward (1.00)

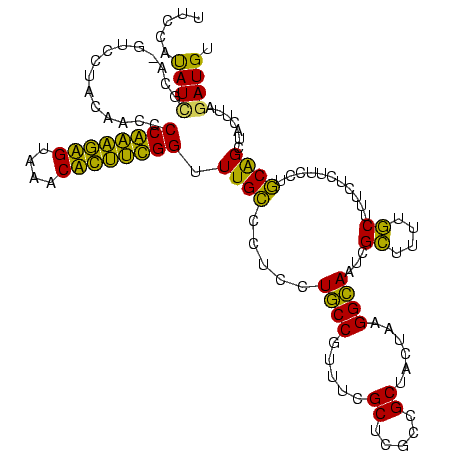
| Location | 580,088 – 580,208 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.96 |
| Mean single sequence MFE | -36.67 |
| Consensus MFE | -29.96 |
| Energy contribution | -25.78 |
| Covariance contribution | -4.19 |
| Combinations/Pair | 1.54 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.82 |
| SVM decision value | 4.14 |
| SVM RNA-class probability | 0.999811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 580088 120 + 2809430 CCUUAGUAGCGGCGAGCGAAAUGGGAAGAGCCCAAACCAACAAGCUUGCUUGUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGACGACAUUAGACGAAUCAUCUGGAAAGAUGA ((((.(((((..(((((....((((.....))))..((((((((....)))))))).)))(((((....))))).))..))))).))))...............((((((....)))))) ( -39.10) >sp_agal.rRNA 18470 113 + 2160275 CCUUAGUAGCGGCGAGCGAAACGGCAGGAGGGCAAACCGAAGUGUUUACACUUCGGGGUUGUAGGAC-UGCGAUAUAGGAUUAAUCAUUA------UAGAAGAAUUACCUGGGAAGGUAA (((.....((.....))......((((....((((.((((((((....))))))))..))))....)-))).....)))...........------........((((((....)))))) ( -33.50) >sp_mutans.rRNA 19051 113 + 2030929 CCUCAGUAGCGGCGAGCGAAGAGGCAGGAGGGCAAACCAGAGUGUUUACACUCUGGGGUUGUAGGAC-UGCGAUAAAGCAGCCAAGGGAA------UAGAAGAAGACUCUGGGAAGAGUC ((((.((..(..(....)..)..))..))))((((.((((((((....))))))))..)))).((.(-(((......)))))).......------........((((((....)))))) ( -41.10) >sp_pneumo.rRNA 17181 113 + 2038623 CCUUAGUAGCGGCGAGCGAAACGGCAGGAGGGCAAACCGAAGAGUUUACUCUUCGGGGUUGUAGGAC-UGCAAUGUGGACUCAAAGAUUA------UAGAAGAAUGAUUUGGGAAGAUCA (((..........((((.(....((((....((((.((((((((....))))))))..))))....)-)))....).).)))..((((((------(......))))))))))....... ( -33.00) >consensus CCUUAGUAGCGGCGAGCGAAACGGCAGGAGGGCAAACCAAAGAGUUUACACUUCGGGGUUGUAGGAC_UGCGAUAUAGAAUCAAAGAGUA______UAGAAGAAUAAUCUGGGAAGAUAA (((.....(((((..........((......))...((((((((....)))))))).)))))))).......................................((((((....)))))) (-29.96 = -25.78 + -4.19)

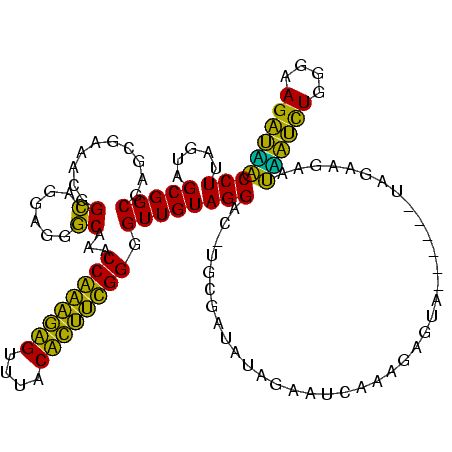
| Location | 580,088 – 580,208 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.96 |
| Mean single sequence MFE | -27.65 |
| Consensus MFE | -22.70 |
| Energy contribution | -18.95 |
| Covariance contribution | -3.75 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.82 |
| SVM decision value | 3.09 |
| SVM RNA-class probability | 0.998401 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 580088 120 - 2809430 UCAUCUUUCCAGAUGAUUCGUCUAAUGUCGUCCUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAACAAGCAAGCUUGUUGGUUUGGGCUCUUCCCAUUUCGCUCGCCGCUACUAAGG ((((((....))))))..((.(....).)).((((.(((....((....(((((..........((((((((....))))))))..((((.....))))...)))))..)).))).)))) ( -29.90) >sp_agal.rRNA 18470 113 - 2160275 UUACCUUCCCAGGUAAUUCUUCUA------UAAUGAUUAAUCCUAUAUCGCA-GUCCUACAACCCCGAAGUGUAAACACUUCGGUUUGCCCUCCUGCCGUUUCGCUCGCCGCUACUAAGG ((((((....))))))........------...........(((.....(((-(.....(((..((((((((....)))))))).))).....))))......((.....)).....))) ( -24.40) >sp_mutans.rRNA 19051 113 - 2030929 GACUCUUCCCAGAGUCUUCUUCUA------UUCCCUUGGCUGCUUUAUCGCA-GUCCUACAACCCCAGAGUGUAAACACUCUGGUUUGCCCUCCUGCCUCUUCGCUCGCCGCUACUGAGG ((((((....))))))........------.......((((((......)))-)))........((((((((....))))))))............((((...((.....))....)))) ( -34.00) >sp_pneumo.rRNA 17181 113 - 2038623 UGAUCUUCCCAAAUCAUUCUUCUA------UAAUCUUUGAGUCCACAUUGCA-GUCCUACAACCCCGAAGAGUAAACUCUUCGGUUUGCCCUCCUGCCGUUUCGCUCGCCGCUACUAAGG ((((........))))........------........((((....((.(((-(.....(((..((((((((....)))))))).))).....)))).))...)))).((........)) ( -22.30) >consensus UAAUCUUCCCAGAUAAUUCUUCUA______UAAUCUUUAAUUCCAUAUCGCA_GUCCUACAACCCCAAAGAGUAAACACUUCGGUUUGCCCUCCUGCCGUUUCGCUCGCCGCUACUAAGG ((((((....)))))).......................................(((.(((..((((((((....)))))))).)))...............((.....)).....))) (-22.70 = -18.95 + -3.75) # Strand winner: forward (1.00)

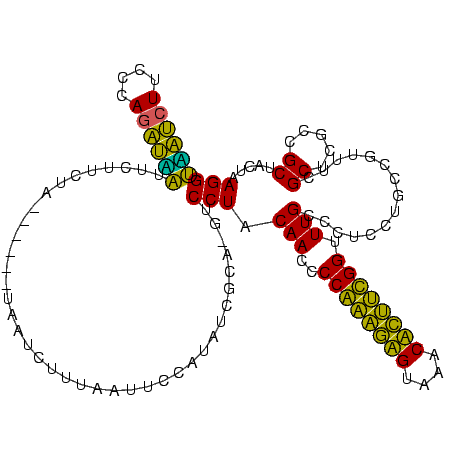
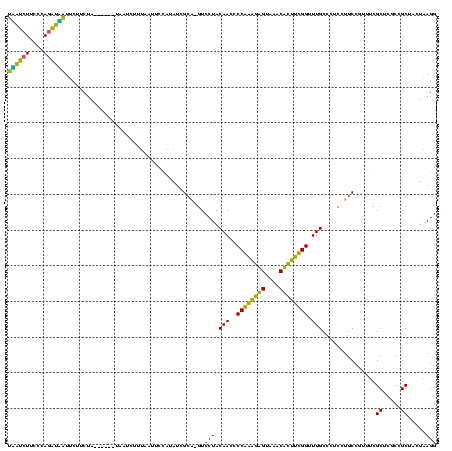
| Location | 580,128 – 580,246 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.67 |
| Mean single sequence MFE | -29.42 |
| Consensus MFE | -21.79 |
| Energy contribution | -18.35 |
| Covariance contribution | -3.44 |
| Combinations/Pair | 1.78 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.86 |
| SVM RNA-class probability | 0.997466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 580128 118 + 2809430 CAAGCUUGCUUGUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGACGACAUUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCGAAAA--UGUUGU ((((....))))......(((((....(((((....))))))))))...((((((((.(((((.((((((....)))))).)).....(((.......)))...)))...))--)))))) ( -30.70) >sp_agal.rRNA 18510 113 + 2160275 AGUGUUUACACUUCGGGGUUGUAGGAC-UGCGAUAUAGGAUUAAUCAUUA------UAGAAGAAUUACCUGGGAAGGUAAGCCAAAGAGAGUAACAGCCUCGUAUUUAAAAUGAUAGAUU ((((....))))(((.((((....)))-).))).....((((.((((((.------((((.(..((((((....))))))..)...(((.((....)))))...)))).)))))).)))) ( -25.90) >sp_mutans.rRNA 19091 111 + 2030929 AGUGUUUACACUCUGGGGUUGUAGGAC-UGCGAUAAAGCAGCCAAGGGAA------UAGAAGAAGACUCUGGGAAGAGUCGCCAGAGAGAGUAAGAGCCUCGUAUUUGAAAU--UCACUU (((((((((.((((..(((....((.(-(((......)))))).......------........((((((....)))))))))..)))).)))))....(((....)))...--.)))). ( -32.30) >sp_pneumo.rRNA 17221 111 + 2038623 AGAGUUUACUCUUCGGGGUUGUAGGAC-UGCAAUGUGGACUCAAAGAUUA------UAGAAGAAUGAUUUGGGAAGAUCAGCCAAAGAGAGUAAUAGCCUCGUAUUUAAAAU--AGUCUU .((((((((((....))((((((....-)))))))))))))).(((((((------(....(..((((((....))))))..)...(((.((....))))).........))--)))))) ( -28.80) >consensus AGAGUUUACACUUCGGGGUUGUAGGAC_UGCGAUAUAGAAUCAAAGAGUA______UAGAAGAAUAAUCUGGGAAGAUAAGCCAAAGAGAGUAAUAGCCUCGUAUUUAAAAU__UGUCUU ((((....)))).(((((((((......(((......))).....................(..((((((....))))))..)..........))))))))).................. (-21.79 = -18.35 + -3.44)

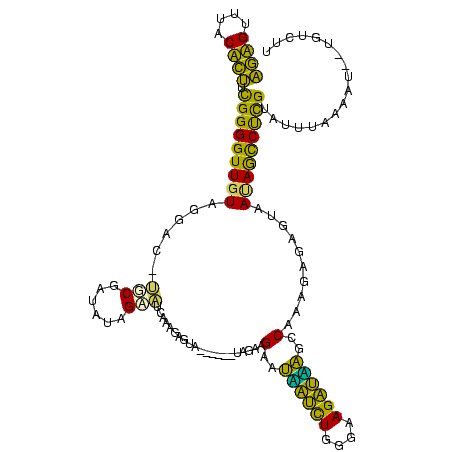

| Location | 580,128 – 580,246 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.67 |
| Mean single sequence MFE | -19.94 |
| Consensus MFE | -13.66 |
| Energy contribution | -9.85 |
| Covariance contribution | -3.81 |
| Combinations/Pair | 1.80 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 580128 118 - 2809430 ACAACA--UUUUCGACUACAGGAUUAUUACCUUCUUUGAUUCAUCUUUCCAGAUGAUUCGUCUAAUGUCGUCCUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAACAAGCAAGCUUG ((.(((--((...(((...(((.......))).....((.((((((....)))))).))))).))))).))...(((((((((......))))....)))))......((((....)))) ( -20.60) >sp_agal.rRNA 18510 113 - 2160275 AAUCUAUCAUUUUAAAUACGAGGCUGUUACUCUCUUUGGCUUACCUUCCCAGGUAAUUCUUCUA------UAAUGAUUAAUCCUAUAUCGCA-GUCCUACAACCCCGAAGUGUAAACACU .......((((((.......(((((((..........(..((((((....))))))..).....------....(((.........))))))-).)))........))))))........ ( -15.26) >sp_mutans.rRNA 19091 111 - 2030929 AAGUGA--AUUUCAAAUACGAGGCUCUUACUCUCUCUGGCGACUCUUCCCAGAGUCUUCUUCUA------UUCCCUUGGCUGCUUUAUCGCA-GUCCUACAACCCCAGAGUGUAAACACU .((((.--.((((......))))...((((...((((((.((((((....))))))........------.......((((((......)))-)))........)))))).)))).)))) ( -30.30) >sp_pneumo.rRNA 17221 111 - 2038623 AAGACU--AUUUUAAAUACGAGGCUAUUACUCUCUUUGGCUGAUCUUCCCAAAUCAUUCUUCUA------UAAUCUUUGAGUCCACAUUGCA-GUCCUACAACCCCGAAGAGUAAACUCU ......--...........(((..((.....((((((((.((((........))))........------......(((((..(........-)..)).)))..))))))))))..))). ( -13.60) >consensus AAGACA__AUUUCAAAUACGAGGCUAUUACUCUCUUUGGCUAAUCUUCCCAGAUAAUUCUUCUA______UAAUCUUUAAUUCCAUAUCGCA_GUCCUACAACCCCAAAGAGUAAACACU ...................(((.......))).....((.((((((....)))))).)).................................................((((....)))) (-13.66 = -9.85 + -3.81) # Strand winner: forward (1.00)

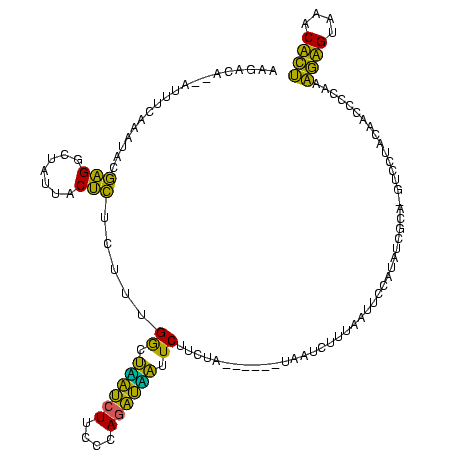
| Location | 580,168 – 580,286 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.67 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -17.48 |
| Energy contribution | -14.72 |
| Covariance contribution | -2.75 |
| Combinations/Pair | 1.63 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
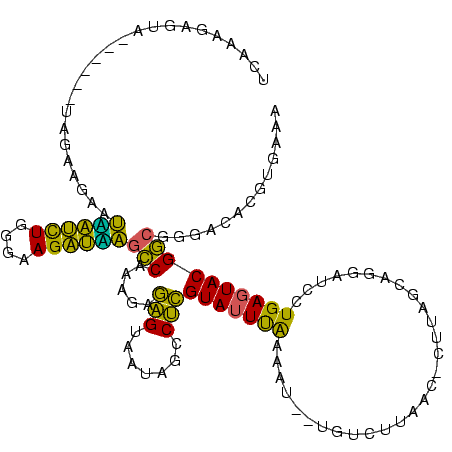
>sy_aureus.rRNA 580168 118 + 2809430 UUACAAAGGACGACAUUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCGAAAA--UGUUGUCUCUCUUGAGUGGAUCCUGAGUACGACGGAGCACGUGAAA ((((...((((((((((.(((((.((((((....)))))).)).....(((.......)))...)))...))--)))))))).......(((..(((..........))).))))))).. ( -34.20) >sp_agal.rRNA 18549 113 + 2160275 UUAAUCAUUA------UAGAAGAAUUACCUGGGAAGGUAAGCCAAAGAGAGUAACAGCCUCGUAUUUAAAAUGAUAGAUUAAC-CUUAGCAGUAUCCUGAGUACGGCGAGACACGCGAAA ...((((((.------((((.(..((((((....))))))..)...(((.((....)))))...)))).))))))........-(((((.......)))))....(((.....))).... ( -21.00) >sp_mutans.rRNA 19130 112 + 2030929 GCCAAGGGAA------UAGAAGAAGACUCUGGGAAGAGUCGCCAGAGAGAGUAAGAGCCUCGUAUUUGAAAU--UCACUUGAUGCCAAGCAGGAUCCUGAGUACGGCGGGACACGAGGAA .((.......------........((((((....))))))(((...(((.((....)))))(((((((..((--((.((((....))))..))))..))))))))))))........... ( -29.60) >sp_pneumo.rRNA 17260 112 + 2038623 UCAAAGAUUA------UAGAAGAAUGAUUUGGGAAGAUCAGCCAAAGAGAGUAAUAGCCUCGUAUUUAAAAU--AGUCUUUGUACUUAGCAGUAUCCUGAGUACGGCGGGACACGUGAAA ..((((((((------(....(..((((((....))))))..)...(((.((....))))).........))--)))))))((((((((.......)))))))).(((.....))).... ( -26.00) >consensus UCAAAGAGUA______UAGAAGAAUAAUCUGGGAAGAUAAGCCAAAGAGAGUAAUAGCCUCGUAUUUAAAAU__UGUCUUAAC_CUUAGCAGGAUCCUGAGUACGGCGGGACACGUGAAA ........................((((((....))))))(((.....(((.......)))(((((((.............................))))))))))............. (-17.48 = -14.72 + -2.75)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Fri Apr 7 15:50:40 2006