
| Sequence ID | sy_sapro.rRNA |
|---|---|
| Location | 748,037 – 748,317 |
| Length | 280 |
| Max. P | 0.990607 |

| Location | 748,037 – 748,157 |
|---|---|
| Length | 120 |
| Sequences | 2 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -52.50 |
| Consensus MFE | -52.50 |
| Energy contribution | -52.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.55 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 748037 120 + 2516583 AGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUA ..(((.((((((((...((.......))...))))))))..)))(((..((((......))))((((((((((((((....))))))).)))))(((((.....))))).....))))). ( -52.50) >sy_haemo.rRNA 884207 120 + 2685023 AGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUA ..(((.((((((((...((.......))...))))))))..)))(((..((((......))))((((((((((((((....))))))).)))))(((((.....))))).....))))). ( -52.50) >consensus AGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUA ..(((.((((((((...((.......))...))))))))..)))(((..((((......))))((((((((((((((....))))))).)))))(((((.....))))).....))))). (-52.50 = -52.50 + 0.00)

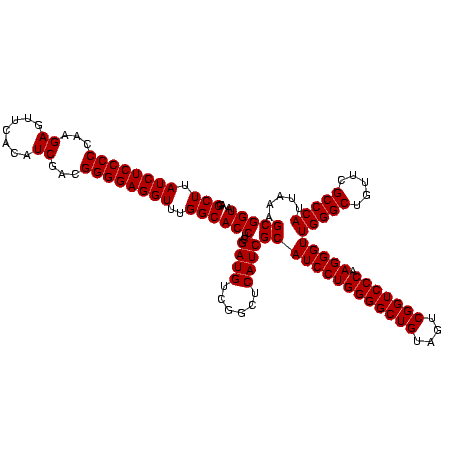
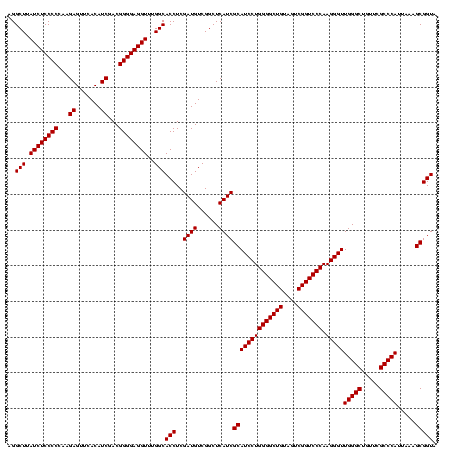
| Location | 748,077 – 748,197 |
|---|---|
| Length | 120 |
| Sequences | 2 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -57.20 |
| Consensus MFE | -54.20 |
| Energy contribution | -57.20 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 748077 120 + 2516583 UGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCC .((.(((..((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..)).(((((((....))).))))))))). ( -57.20) >sy_haemo.rRNA 884247 120 + 2685023 UGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCC .((.(((..((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..)).(((((((....))).))))))))). ( -57.20) >consensus UGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCC .((.(((..((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..)).(((((((....))).))))))))). (-54.20 = -57.20 + 3.00)

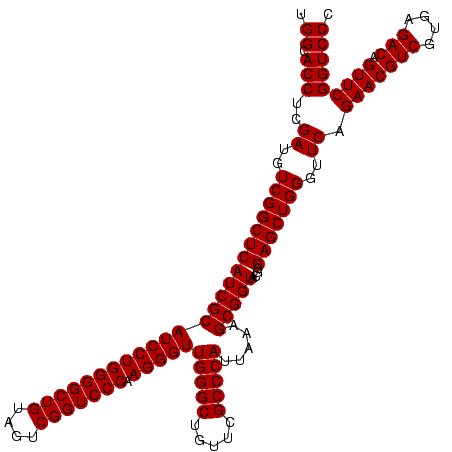
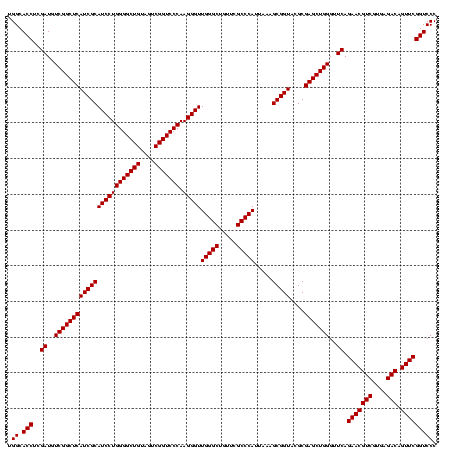
| Location | 748,197 – 748,317 |
|---|---|
| Length | 120 |
| Sequences | 2 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -43.60 |
| Consensus MFE | -41.60 |
| Energy contribution | -41.60 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.95 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.505808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 748197 120 + 2516583 UAUCCGUCGUGGGCGUAGGAAAUUUGAGAGGAGCUGUCCUUAGUACGAGAGGACCGGGAUGGACAUACCUCUGGUGUACCAGUUGUCGUGCCAACGGCAUCGCUGGGUAGCUAUGUAUGG ..((((((.(((((..((((..(((....)))....))))..)).(....)..))).))))))(((((...((((...(((((((((((....))))))..)))))...)))).))))). ( -43.60) >sy_haemo.rRNA 884367 120 + 2685023 UAUCCGUCGUGGGCGUAGGAAAUUUGAGAGGAGCUGUCCUUAGUACGAGAGGACCGGGAUGGACAUACCUCUGGUGUACCAGUUGUCGUGCCAACGGCAUAGCUGGGUAGCUAUGUAUGG ..((((((.(((((..((((..(((....)))....))))..)).(....)..))).))))))(((((...((((...(((((((((((....))))))..)))))...)))).))))). ( -43.60) >consensus UAUCCGUCGUGGGCGUAGGAAAUUUGAGAGGAGCUGUCCUUAGUACGAGAGGACCGGGAUGGACAUACCUCUGGUGUACCAGUUGUCGUGCCAACGGCAUAGCUGGGUAGCUAUGUAUGG ..((((((.(((...............(((((....)))))....(....)..))).))))))(((((...((((...(((((((((((....))))))..)))))...)))).))))). (-41.60 = -41.60 + 0.00)

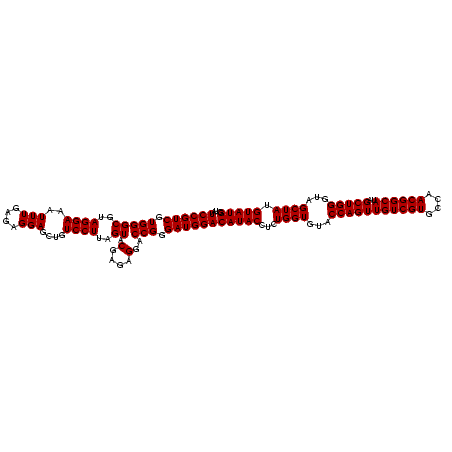
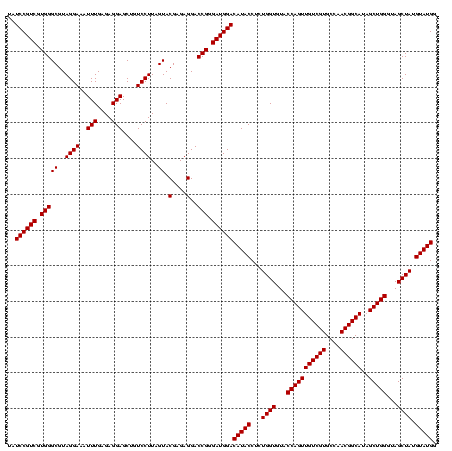
| Location | 748,197 – 748,317 |
|---|---|
| Length | 120 |
| Sequences | 2 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -31.55 |
| Consensus MFE | -31.60 |
| Energy contribution | -32.10 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.849101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 748197 120 - 2516583 CCAUACAUAGCUACCCAGCGAUGCCGUUGGCACGACAACUGGUACACCAGAGGUAUGUCCAUCCCGGUCCUCUCGUACUAAGGACAGCUCCUCUCAAAUUUCCUACGCCCACGACGGAUA .....(((.(((....))).)))((((((((........(((....)))((((..(((((....(((.....)))......)))))...)))).............)))...)))))... ( -29.30) >sy_haemo.rRNA 884367 120 - 2685023 CCAUACAUAGCUACCCAGCUAUGCCGUUGGCACGACAACUGGUACACCAGAGGUAUGUCCAUCCCGGUCCUCUCGUACUAAGGACAGCUCCUCUCAAAUUUCCUACGCCCACGACGGAUA .....(((((((....)))))))((((((((........(((....)))((((..(((((....(((.....)))......)))))...)))).............)))...)))))... ( -33.80) >consensus CCAUACAUAGCUACCCAGCGAUGCCGUUGGCACGACAACUGGUACACCAGAGGUAUGUCCAUCCCGGUCCUCUCGUACUAAGGACAGCUCCUCUCAAAUUUCCUACGCCCACGACGGAUA .....(((((((....)))))))((((((((........(((....)))((((..(((((....(((.....)))......)))))...)))).............)))...)))))... (-31.60 = -32.10 + 0.50) # Strand winner: forward (1.00)

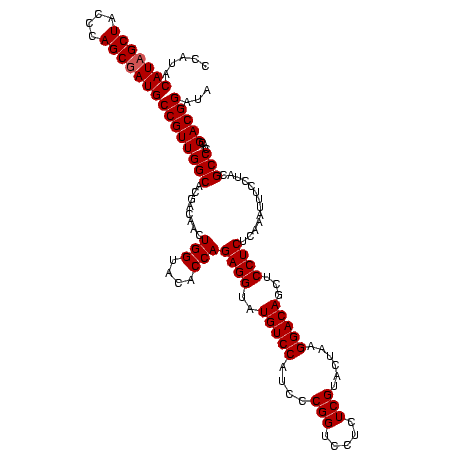
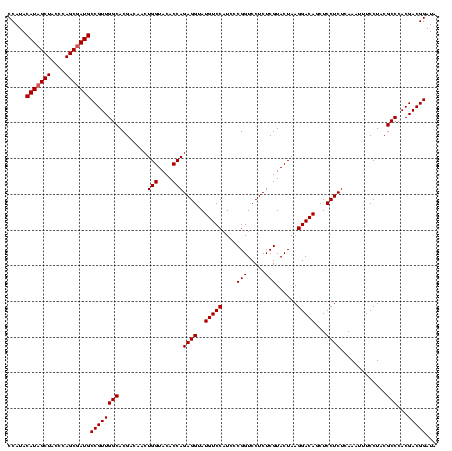
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Fri Apr 7 15:51:52 2006