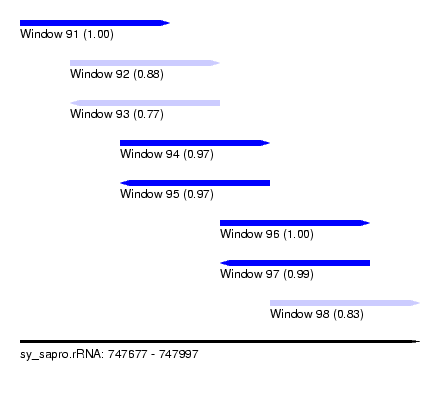
| Sequence ID | sy_sapro.rRNA |
|---|---|
| Location | 747,677 – 747,997 |
| Length | 320 |
| Max. P | 0.999388 |

| Location | 747,677 – 747,797 |
|---|---|
| Length | 120 |
| Sequences | 2 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -47.60 |
| Consensus MFE | -45.85 |
| Energy contribution | -46.85 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.96 |
| SVM RNA-class probability | 0.997931 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 747677 120 + 2516583 GAAAUUCGGCACAGCUUGUACAGGAUAGGUAGGAGCCUGAGAUACGUGAGCGCUAGCUUACGUGGAGGCGUUGGUGGGAUACUACCCUCGCUGUGUUGGAUUUCUAACCCGCACCAUUUA (((((((((((((((((.(((.......))).))((((....(((((((((....))))))))).))))(..(((((....)))))..))))))))))))))))................ ( -49.70) >sy_haemo.rRNA 883847 120 + 2685023 GAAAUUCGGCACAGCUUGUACAGGAUAGGUAGGAGCCUUUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUA ((((..((((((((((.(.............((.(((((....((((((((....)))))))).))))).))(((((....)))))).))))))))))..))))................ ( -45.50) >consensus GAAAUUCGGCACAGCUUGUACAGGAUAGGUAGGAGCCUGAGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGAUUUCUAACCCGCACCACUUA (((((((((((((((((.(((.......))).))((((....(((((((((....))))))))).))))...(((((....)))))...)))))))))))))))................ (-45.85 = -46.85 + 1.00)

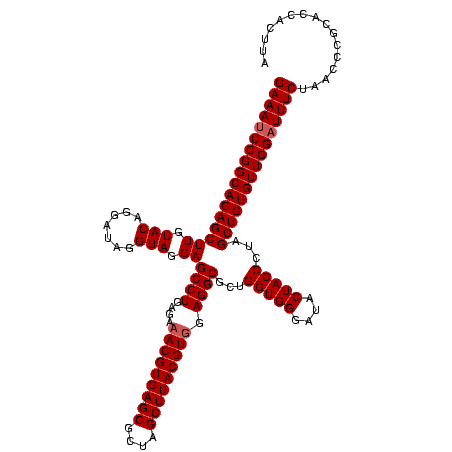

| Location | 747,717 – 747,837 |
|---|---|
| Length | 120 |
| Sequences | 2 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -46.95 |
| Consensus MFE | -44.90 |
| Energy contribution | -43.90 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.879567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 747717 120 + 2516583 GAUACGUGAGCGCUAGCUUACGUGGAGGCGUUGGUGGGAUACUACCCUCGCUGUGUUGGAUUUCUAACCCGCACCAUUUAUCAUGGUGGGAGACAGUGUCAGAUGGGCAGUUUGACUGGG ..(((((((((....)))))))))..((((..(((((....)))))..))))..(((((....)))))((.((((((.....))))))(....)...(((((((.....))))))).)). ( -46.20) >sy_haemo.rRNA 883887 120 + 2685023 GAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGGCGGGCAGUUUGACUGGG ...((((((((....))))))))(((((((((((.((.......))))))).......))))))...(((.((((((.....))))))(....)...(((((((.....))))))).))) ( -47.71) >consensus GAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGAUUUCUAACCCGCACCACUUAUCAUGGUGGGAGACAGUGUCAGACGGGCAGUUUGACUGGG ...((((((((....)))))))).........(((((....)))))...((((((((((....)))))((.((((((.....))))))))..)))))(((((((.....))))))).... (-44.90 = -43.90 + -1.00)

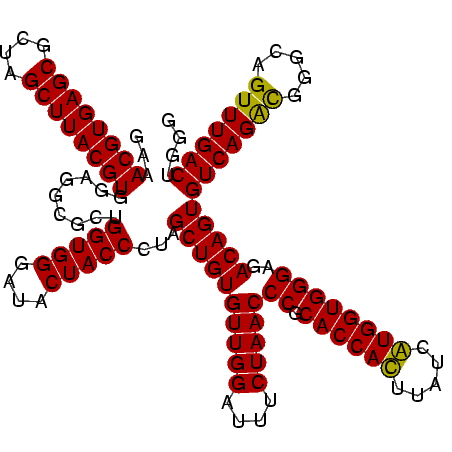
| Location | 747,717 – 747,837 |
|---|---|
| Length | 120 |
| Sequences | 2 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -34.10 |
| Consensus MFE | -33.00 |
| Energy contribution | -32.50 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 747717 120 - 2516583 CCCAGUCAAACUGCCCAUCUGACACUGUCUCCCACCAUGAUAAAUGGUGCGGGUUAGAAAUCCAACACAGCGAGGGUAGUAUCCCACCAACGCCUCCACGUAAGCUAGCGCUCACGUAUC .........(((((((.((((((.(((.....((((((.....)))))))))))))))..(((......).))))))))).................((((.(((....))).))))... ( -32.90) >sy_haemo.rRNA 883887 120 - 2685023 CCCAGUCAAACUGCCCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAGCGCUCACGUUUC .........(((((((..(((((.(((.....((((((.....))))))))))))))..(((.......))).)))))))........(((((.....(....)...)))))........ ( -35.30) >consensus CCCAGUCAAACUGCCCACCUGACACUGUCUCCCACCACGAUAAAUGGUGCGGGUUAGAAAGCCAACACAGCGAGGGUAGUAUCCCACCAACGCCUCCACGUAAGCUAGCGCUCACGUAUC .........(((((((..(((.........((((((((.....)))))).))(((.(....).))).)))...))))))).................((((.(((....))).))))... (-33.00 = -32.50 + -0.50) # Strand winner: forward (1.00)

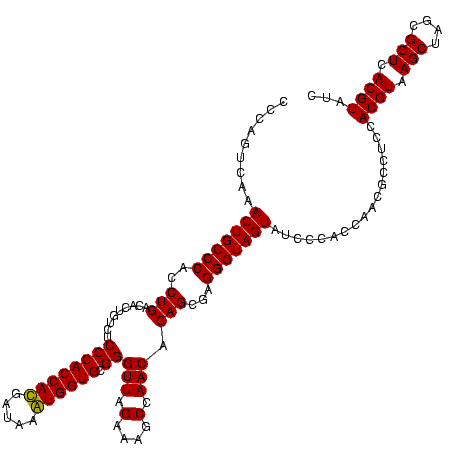
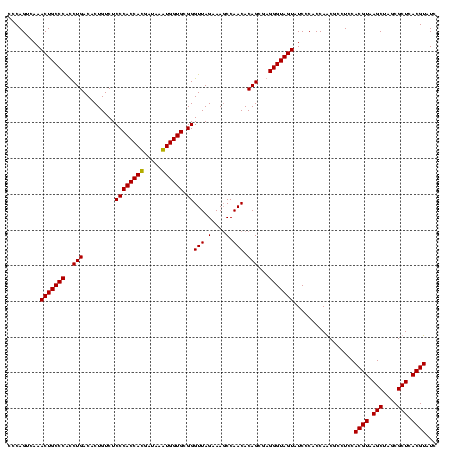
| Location | 747,757 – 747,877 |
|---|---|
| Length | 120 |
| Sequences | 2 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -46.35 |
| Consensus MFE | -46.85 |
| Energy contribution | -45.85 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.76 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 747757 120 + 2516583 ACUACCCUCGCUGUGUUGGAUUUCUAACCCGCACCAUUUAUCAUGGUGGGAGACAGUGUCAGAUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGAGUAACGGAGGCGCUCAAAGGUU ((..((((.((((((((((....)))))((.((((((.....))))))))..)))))(((((((.....))))))).))))..))(((((((...........))))))).......... ( -44.30) >sy_haemo.rRNA 883927 120 + 2685023 ACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGGCGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUU ((..(((((((((((((((....)))))((.((((((.....))))))))..)))))(((((((.....))))))))))))..))(((((((...........))))))).......... ( -48.40) >consensus ACUACCCUAGCUGUGUUGGAUUUCUAACCCGCACCACUUAUCAUGGUGGGAGACAGUGUCAGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAAGUAACGGAGGCGCUCAAAGGUU ((..((((.((((((((((....)))))((.((((((.....))))))))..)))))(((((((.....))))))).))))..))(((((((...........))))))).......... (-46.85 = -45.85 + -1.00)

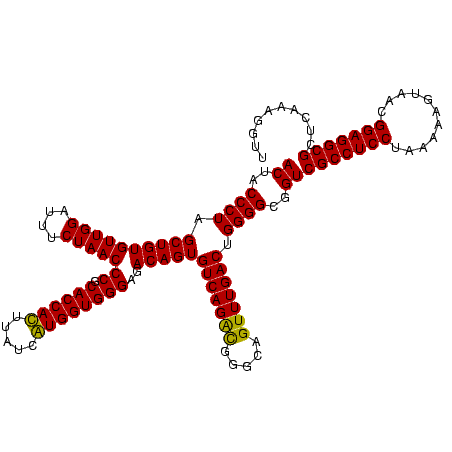

| Location | 747,757 – 747,877 |
|---|---|
| Length | 120 |
| Sequences | 2 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -41.10 |
| Consensus MFE | -40.10 |
| Energy contribution | -39.60 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 747757 120 - 2516583 AACCUUUGAGCGCCUCCGUUACUCUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCAUCUGACACUGUCUCCCACCAUGAUAAAUGGUGCGGGUUAGAAAUCCAACACAGCGAGGGUAGU .......(..(((((((...........)))))))..)...........(((((((.((((((.(((.....((((((.....)))))))))))))))..(((......).))))))))) ( -39.70) >sy_haemo.rRNA 883927 120 - 2685023 AACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAGCUAGGGUAGU .......(..(((((((...........)))))))..)...........(((((((..(((((.(((.....((((((.....))))))))))))))..(((.......))).))))))) ( -42.50) >consensus AACCUUUGAGCGCCUCCGUUACCCUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCACCACGAUAAAUGGUGCGGGUUAGAAAGCCAACACAGCGAGGGUAGU .......(..(((((((...........)))))))..)...........(((((((..(((.........((((((((.....)))))).))(((.(....).))).)))...))))))) (-40.10 = -39.60 + -0.50) # Strand winner: forward (0.99)

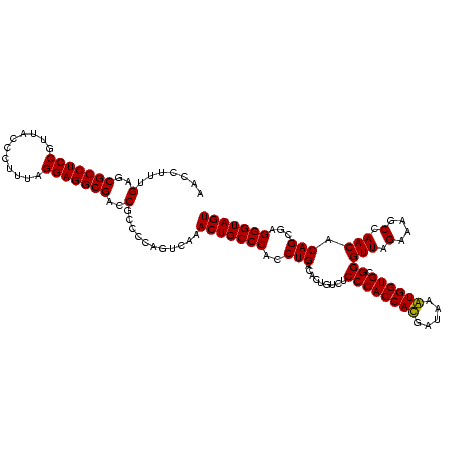
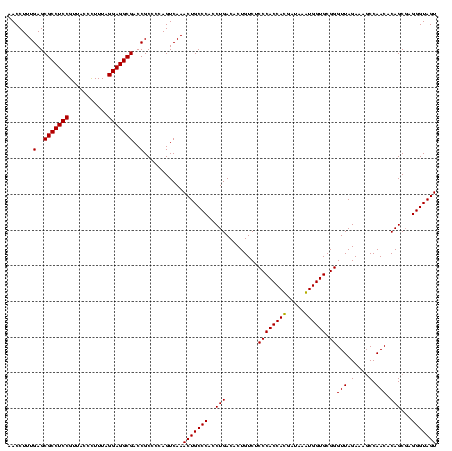
| Location | 747,837 – 747,957 |
|---|---|
| Length | 120 |
| Sequences | 2 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -44.10 |
| Consensus MFE | -44.35 |
| Energy contribution | -43.85 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.31 |
| Structure conservation index | 1.01 |
| SVM decision value | 3.56 |
| SVM RNA-class probability | 0.999388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 747837 120 + 2516583 GCGGUCGCCUCCUAAAGAGUAACGGAGGCGCUCAAAGGUUUCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGAAGCUUGACUGCGAGACUUACAAGUCGAGCAGGG ..(..(((((((...........)))))))..)..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..((((....))))..)))).. ( -43.50) >sy_haemo.rRNA 884007 120 + 2685023 GCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGG ..(..(((((((...........)))))))..)..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).. ( -44.70) >consensus GCGGUCGCCUCCUAAAAAGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGAAGCUUGACUGCGAGACCUACAAGUCGAGCAGGG ..(..(((((((...........)))))))..)..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).. (-44.35 = -43.85 + -0.50)

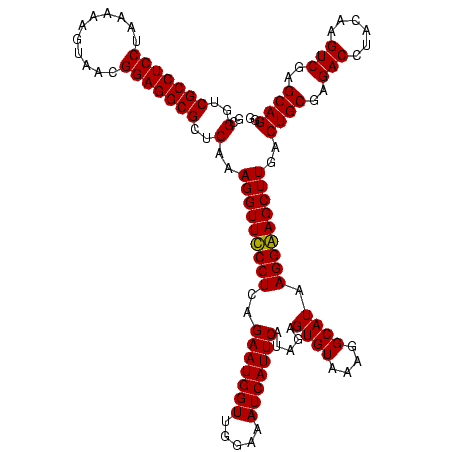
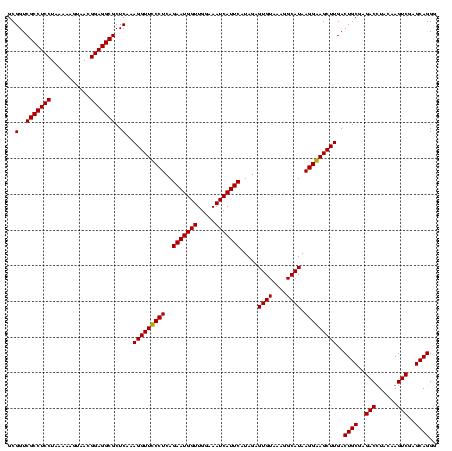
| Location | 747,837 – 747,957 |
|---|---|
| Length | 120 |
| Sequences | 2 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -34.12 |
| Consensus MFE | -34.30 |
| Energy contribution | -33.55 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.15 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 747837 120 - 2516583 CCCUGCUCGACUUGUAAGUCUCGCAGUCAAGCUUCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGAAACCUUUGAGCGCCUCCGUUACUCUUUAGGAGGCGACCGC ..((((..((((....))))..))))(((((.(((((((((......))......(((((.........))))))))))))...))))).(((((((...........)))))))..... ( -32.20) >sy_haemo.rRNA 884007 120 - 2685023 CCCUGCUCGACUUGUAGGUCUCGCAGUCAAGCUCCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGC .(((((.......)))))....((.(((..((((.......((((((........(((((.........))))))))))).......))))((((((...........))))))))).)) ( -36.04) >consensus CCCUGCUCGACUUGUAAGUCUCGCAGUCAAGCUCCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGAAACCUUUGAGCGCCUCCGUUACCCUUUAGGAGGCGACCGC ..((((..((((....))))..))))(((((.(((((((((......))......(((((.........))))))))))))...))))).(((((((...........)))))))..... (-34.30 = -33.55 + -0.75) # Strand winner: forward (1.00)

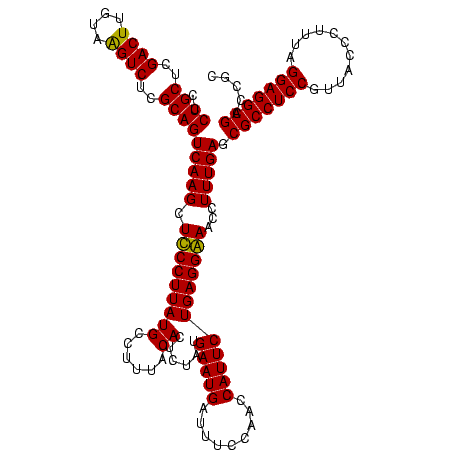

| Location | 747,877 – 747,997 |
|---|---|
| Length | 120 |
| Sequences | 2 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -36.70 |
| Consensus MFE | -36.85 |
| Energy contribution | -36.35 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.55 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 747877 120 + 2516583 UCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGAAGCUUGACUGCGAGACUUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUGAUCCGGUGGUUCCGCAUGGAA (((.(.((((.(..(((......((((.((((.(..((((........))))..((((..((((....))))..)))).(((....)))).)))).)))))))..).)))).)...))). ( -36.20) >sy_haemo.rRNA 884047 120 + 2685023 CCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUGAUCCGGUGGUUCCGCAUGGAA ((((..(((((((.....))))))).....((((....)))).)))).((((((((............))))))))...(((....)))...........(((((((....))).)))). ( -37.20) >consensus CCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGAAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUGAUCCGGUGGUUCCGCAUGGAA ((((..(((((((.....))))))).....((((....)))).)))).((((((((............))))))))...(((....)))...........(((((((....))).)))). (-36.85 = -36.35 + -0.50)


Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Fri Apr 7 15:51:48 2006