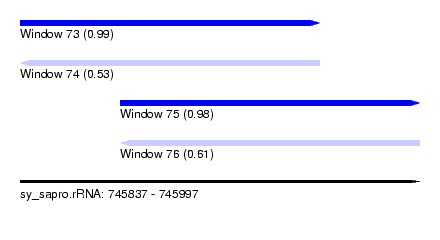
| Sequence ID | sy_sapro.rRNA |
|---|---|
| Location | 745,837 – 745,997 |
| Length | 160 |
| Max. P | 0.994699 |

| Location | 745,837 – 745,957 |
|---|---|
| Length | 120 |
| Sequences | 2 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.17 |
| Mean single sequence MFE | -29.50 |
| Consensus MFE | -30.15 |
| Energy contribution | -29.65 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.89 |
| Structure conservation index | 1.02 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
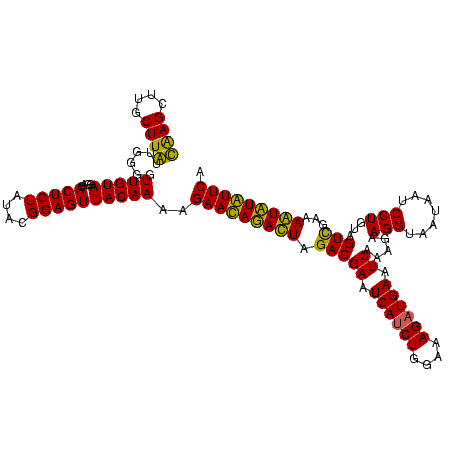
>sy_sapro.rRNA 745837 120 + 2516583 CGAGCUUGCUUGUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAAGAACAGACUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCGAAAGUUUGUUCA ((((....))))......(((((....(((((....))))))))))..(((((((((.(((((.((((((....)))))).)).....(((.......)))...)))...))))))))). ( -35.00) >sy_haemo.rRNA 882008 120 + 2685023 UAAGCUUGCUUAUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGAAUAUAUUAGACGAAUCAUCUGGAAAGUUGAAUCAAAGAAGGUAAUAAUCCUGUAGUUGAAAAUAUAUUCU ((((....))))......(((((....(((((....)))))))))).((((((((((.(((((.(((.((....)).))).)).....(((.......)))...)))...)))))))))) ( -24.00) >consensus CAAGCUUGCUUAUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAAGAACAGACUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCGAAAAUAUAUUCA ((((....))))......(((((....(((((....))))))))))..(((((((((.(((((.((((((....)))))).)).....(((.......)))...)))...))))))))). (-30.15 = -29.65 + -0.50)

| Location | 745,837 – 745,957 |
|---|---|
| Length | 120 |
| Sequences | 2 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.17 |
| Mean single sequence MFE | -19.50 |
| Consensus MFE | -19.55 |
| Energy contribution | -20.05 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.08 |
| Mean z-score | -0.91 |
| Structure conservation index | 1.00 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
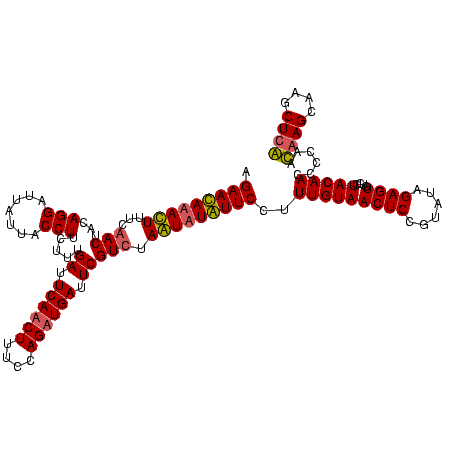
>sy_sapro.rRNA 745837 120 - 2516583 UGAACAAACUUUCGACUACAGGAUUAUUACCUUCUUUGAUUCAUCUUUCCAGAUGAUUCGUCUAGUCUGUUCUUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAACAAGCAAGCUCG .(((((.(((...(((...(((.......))).....((.((((((....)))))).))))).))).)))))..(((((((((......))))....))))).................. ( -22.60) >sy_haemo.rRNA 882008 120 - 2685023 AGAAUAUAUUUUCAACUACAGGAUUAUUACCUUCUUUGAUUCAACUUUCCAGAUGAUUCGUCUAAUAUAUUCCUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAAUAAGCAAGCUUA .(((((((((..........((((((..........))))))........(((((...))))))))))))))..(((((((((......))))....)))))......((((....)))) ( -16.40) >consensus AGAACAAACUUUCAACUACAGGAUUAUUACCUUCUUUGAUUCAACUUUCCAGAUGAUUCGUCUAAUAUAUUCCUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAACAAGCAAGCUCA .(((((((((...(((...(((.......))).....((.((((((....)))))).))))).)))))))))..(((((((((......))))....)))))......((((....)))) (-19.55 = -20.05 + 0.50) # Strand winner: forward (1.00)

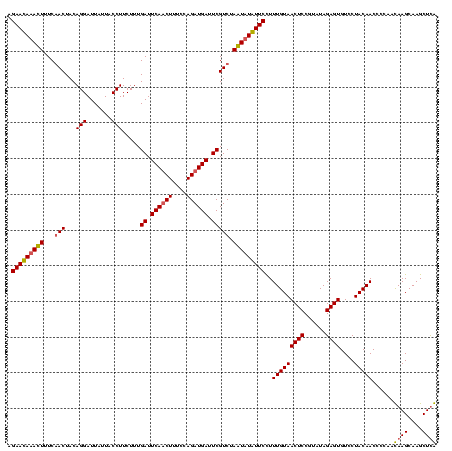
| Location | 745,877 – 745,997 |
|---|---|
| Length | 120 |
| Sequences | 2 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -28.65 |
| Consensus MFE | -27.00 |
| Energy contribution | -27.75 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 745877 120 + 2516583 UUACAAAAGAACAGACUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCGAAAGUUUGUUCACUCUUGAGUGGAUCCUGAGUACGACGGAACACGAGAAAUU ........(((((((((.(((((.((((((....)))))).)).....(((.......)))...)))...)))))))))..((((..(((..(((..........))).))))))).... ( -32.90) >sy_haemo.rRNA 882048 120 + 2685023 UUACAAAGGAAUAUAUUAGACGAAUCAUCUGGAAAGUUGAAUCAAAGAAGGUAAUAAUCCUGUAGUUGAAAAUAUAUUCUCUCUUGAGUGGAUCCUGAGUACGACGGAGCACGUGAAAUU ((((...((((((((((.(((((.(((.((....)).))).)).....(((.......)))...)))...)))))))))).......(((..(((..........))).))))))).... ( -24.40) >consensus UUACAAAAGAACAGACUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCGAAAAUAUAUUCACUCUUGAGUGGAUCCUGAGUACGACGGAACACGAGAAAUU ((((....(((((((((.(((((.((((((....)))))).)).....(((.......)))...)))...)))))))))........(((..(((..........))).))))))).... (-27.00 = -27.75 + 0.75)

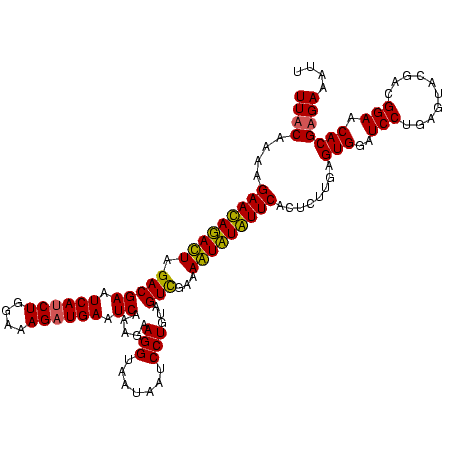
| Location | 745,877 – 745,997 |
|---|---|
| Length | 120 |
| Sequences | 2 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -21.30 |
| Consensus MFE | -21.40 |
| Energy contribution | -21.65 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.01 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sapro.rRNA 745877 120 - 2516583 AAUUUCUCGUGUUCCGUCGUACUCAGGAUCCACUCAAGAGUGAACAAACUUUCGACUACAGGAUUAUUACCUUCUUUGAUUCAUCUUUCCAGAUGAUUCGUCUAGUCUGUUCUUUUGUAA ........(((.(((..........)))..))).(((((..(((((.(((...(((...(((.......))).....((.((((((....)))))).))))).))).))))))))))... ( -24.00) >sy_haemo.rRNA 882048 120 - 2685023 AAUUUCACGUGCUCCGUCGUACUCAGGAUCCACUCAAGAGAGAAUAUAUUUUCAACUACAGGAUUAUUACCUUCUUUGAUUCAACUUUCCAGAUGAUUCGUCUAAUAUAUUCCUUUGUAA ........(((.(((..........)))..)))...((((.(((((((((..........((((((..........))))))........(((((...)))))))))))))))))).... ( -18.60) >consensus AAUUUCACGUGCUCCGUCGUACUCAGGAUCCACUCAAGAGAGAACAAACUUUCAACUACAGGAUUAUUACCUUCUUUGAUUCAACUUUCCAGAUGAUUCGUCUAAUAUAUUCCUUUGUAA ........(((.(((..........)))..)))...((((.(((((((((...(((...(((.......))).....((.((((((....)))))).))))).))))))))))))).... (-21.40 = -21.65 + 0.25) # Strand winner: forward (0.99)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Fri Apr 7 15:51:30 2006