| Sequence ID | sy_sapro.tRNA |
|---|---|
| Location | 1,004,151 – 1,004,208 |
| Length | 57 |
| Max. P | 0.999432 |

| Location | 1,004,151 – 1,004,208 |
|---|---|
| Length | 57 |
| Sequences | 2 |
| Columns | 58 |
| Reading direction | reverse |
| Mean pairwise identity | 82.76 |
| Mean single sequence MFE | -18.50 |
| Consensus MFE | -18.30 |
| Energy contribution | -17.30 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.60 |
| SVM RNA-class probability | 0.999432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
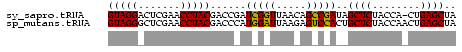
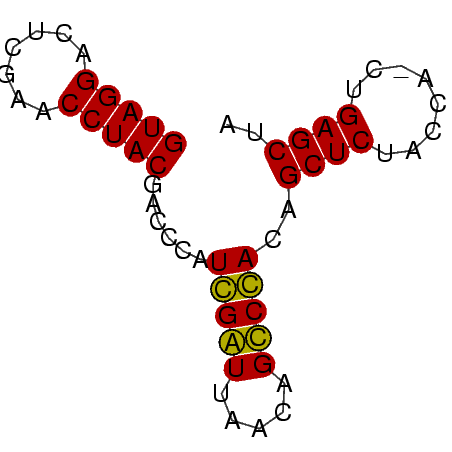
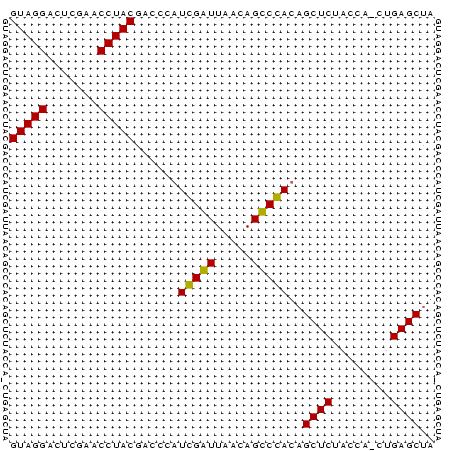
>sy_sapro.tRNA 1004151 57 - 2516583 GUAGGACUCGAACCUACGACCGAUCGGUUAACAGCCGAUAGCUCUACCA-CUGAGCUA (((((.......))))).....((((((.....))))))(((((.....-..))))). ( -19.60) >sp_mutans.tRNA 1928150 58 - 2030929 GUAGGGCUCGAACCUACGACCCAUGGAUUAAGAGUCCACUGCUCUACCAACUGAGCUA ...(((.(((......)))))).(((((.....)))))..((((........)))).. ( -17.40) >consensus GUAGGACUCGAACCUACGACCCAUCGAUUAACAGCCCACAGCUCUACCA_CUGAGCUA (((((.......)))))......(((((.....)))))..((((........)))).. (-18.30 = -17.30 + -1.00) # Strand winner: reverse (0.57)
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Fri Apr 7 15:51:23 2006