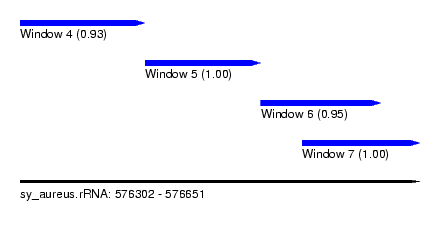
| Sequence ID | sy_aureus.rRNA |
|---|---|
| Location | 576,302 – 576,651 |
| Length | 349 |
| Max. P | 0.999747 |

| Location | 576,302 – 576,411 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 62.05 |
| Mean single sequence MFE | -30.77 |
| Consensus MFE | -15.71 |
| Energy contribution | -17.02 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.934105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 576302 109 + 2809430 AAAAUAGGUGCCCGUACCGCAAACCGACACAGGUAGUCAAGAUGAGAAUUCUAAGGUGA------GCGAGCGAACUCUCG-----UUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGG ........((((.((.((((..((((((.......))).(((.......)))..)))..------)).(((((....)))-----))..)).))..))))........(((......))) ( -24.20) >sp_agal.rRNA 16974 105 + 2160275 ----UUAUUGGGCGUA----AAGCGAGCGCAGGCGGUUC----UUUAAGUCUGAAGUUAAAGGCAGUGGCUUAACCAUUGUACGCUUUGGAAACUGGAGGACUUGAGUGCAGAA----GG ----.......((...----..))..((((....(((((----((((..((..((((.....(((((((.....)))))))..))))..))...)))))))))...))))....----.. ( -31.80) >sp_mutans.rRNA 17448 105 + 2030929 ----UUAUUGGGCGUA----AAGGGAGCGCAGGCGGUCA----GGAAAGUCUGGAGUAAAAGGCUAUGGCUCAACCAUAGUGUGCUCUGGAAACUGUCUGACUUGAGUGCAGAA----GG ----.......((...----......))(((..((((((----((....((..(((((....(((((((.....))))))).)))))..)).....)))))).))..)))....----.. ( -35.20) >sp_pneumo.rRNA 15719 105 + 2038623 ----UUAUUGGGCGUA----AAGCGAGCGCAGGCGGUUA----GAUAAGUCUGAAGUUAAAGGCUGUGGCUUAACCAUAGUAGGCUUUGGAAACUGUUUAACUUGAGUGCAAGA----GG ----.......((...----..))..((((....(((((----((((..((..(((((....(((((((.....))))))).)))))..))...)))))))))...))))....----.. ( -31.90) >consensus ____UUAUUGGGCGUA____AAGCGAGCGCAGGCGGUCA____GAUAAGUCUGAAGUUAAAGGCUGUGGCUUAACCAUAGUA_GCUUUGGAAACUGGAUAACUUGAGUGCAGAA____GG ...........((((...........))))..(((.(((....((((..(((((((((....(((((((.....))))))).)))))))))...)))).....))).))).......... (-15.71 = -17.02 + 1.31)


| Location | 576,411 – 576,512 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.96 |
| Mean single sequence MFE | -37.77 |
| Consensus MFE | -21.69 |
| Energy contribution | -20.88 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.99 |
| SVM RNA-class probability | 0.998055 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 576411 101 + 2809430 GAGAAGGGGUGCUCUUUAGG--------------GUUAACGCCCAGAAGAGCCGCAGUGAAUAGGCCCAAGCGACU-----GUUUAUCAAAAACACAGGUCUCUGCUAAACCGUAAGGUG ((((..(.(.(((((((.((--------------((....)))).)))))))).).((((((((((....))..))-----))))))............))))......(((....))). ( -31.20) >sp_agal.rRNA 17079 112 + 2160275 GGAGAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAUAUAUGGAGGAACACCGGU-----GGCGAAAGCGGCUCUCUGGUCUGUAACUGACGCUGAGGCUCG--AAAGCGU-GGGGA ((((((((...((((((((((.(((......)))....))))))))))...)))..((-----.((....)).))))))).........((.(((((........--..)))))-.)).. ( -39.60) >sp_mutans.rRNA 17553 112 + 2030929 GGAGAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAUAUAUGGAGGAACACCAGU-----GGCGAAAGCGGCUCUCUGGUCUGUCACUGACGCUGAGGCUCG--AAAGCGU-GGGUA ((((((((...((((((((((.(((......)))....))))))))))...)))..((-----.((....)).)))))))........(((.(((((........--..)))))-.))). ( -40.70) >sp_pneumo.rRNA 15824 112 + 2038623 GGAGAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAUAUAUGGAGGAACACCGGU-----GGCGAAAGCGGCUCUCUGGCUUGUAACUGACGCUGAGGCUCG--AAAGCGU-GGGGA ((((((((...((((((((((.(((......)))....))))))))))...)))..((-----.((....)).))))))).........((.(((((........--..)))))-.)).. ( -39.60) >consensus GGAGAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAUAUAUGGAGGAACACCAGU_____GGCGAAAGCGGCUCUCUGGUCUGUAACUGACGCUGAGGCUCG__AAAGCGU_GGGGA .(((.(((...((((((((((.(((......)))....))))))))))...)))((((......((....))(((......)))..........))))...)))................ (-21.69 = -20.88 + -0.81)

| Location | 576,512 – 576,617 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.92 |
| Mean single sequence MFE | -42.33 |
| Consensus MFE | -23.00 |
| Energy contribution | -23.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.54 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 576512 105 + 2809430 AUGUAUAGGGGCUGACGCC----UGCCCGGUGCUGGAAGGUUAAGAGGAGUG------GUUAG--CUUCUGCGAAGCUACGAAUCGAAGC---CCCAGUAAACGGCGGCCGUAACUAUAA ...(((((..((....(((----.(((...((((((..((((..((...(((------(((.(--(....))..))))))...))..)))---)))))))...)))))).))..))))). ( -38.70) >sp_agal.rRNA 17191 116 + 2160275 GCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAA----ACGAUGAGUGCUAGGUGUUAGGCCCUUUCCGGGGCUUAGUGCCGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUA .....((((.........))))....((((.((((...----.))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).)).))))..... ( -44.70) >sp_mutans.rRNA 17665 116 + 2030929 GCGAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAA----ACGAUGAGUGCUAGGUGUUAGGCCCUUUCCGGGGCUUAGUGCCGGAGCUAACGCAAUAAGCACUCCGCCUGGGGAGUA .....((((.........))))....((((.((((...----.))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).)).))))..... ( -44.70) >sp_pneumo.rRNA 15936 116 + 2038623 GCAAACAGGAUUAGAUACCCUGGUAGUCCACGCUGUAA----ACGAUGAGUGCUAGGUGUUAGACCCUUUCCGGGGUUUAGUGCCGUAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUA .................(((.((((((....)))....----.....(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))).)))..... ( -41.20) >consensus GCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAA____ACGAUGAGUGCUAGGUGUUAGGCCCUUUCCGGGGCUUAGUGCCGAAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUA ((.....((((((..........))))))...................(((....((..(((((((((....)))))))))..))...)))..........))(((((......))))). (-23.00 = -23.00 + 0.00)

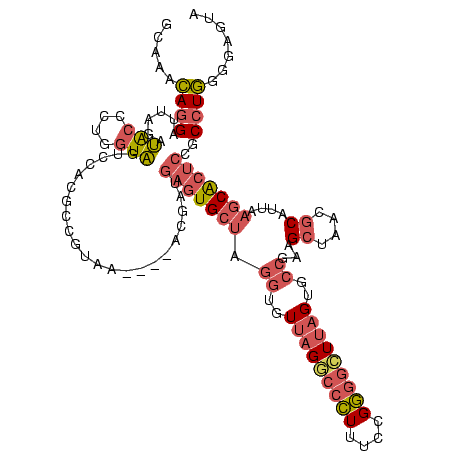

| Location | 576,548 – 576,651 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.39 |
| Mean single sequence MFE | -45.95 |
| Consensus MFE | -23.51 |
| Energy contribution | -25.95 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.28 |
| Mean z-score | -3.46 |
| Structure conservation index | 0.51 |
| SVM decision value | 3.99 |
| SVM RNA-class probability | 0.999747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 576548 103 + 2809430 UUAAGAGGAGUG------GUUAG--CUUCUGCGAAGCUACGAAUCGAAGC---CCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUC------CUUGUCGGGUAA ....(((((((.------..(((--((((...)))))))....(((..((---(((.......)).((((((.......))))))....)))..))).))))------)))......... ( -28.30) >sp_agal.rRNA 17228 114 + 2160275 --ACGAUGAGUGCUAGGUGUUAGGCCCUUUCCGGGGCUUAGUGCCGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGU--UGAAACUCAAAGGAAUUGACGGG--- --.(((((((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))--))..))))..))).)))).....--- ( -52.50) >sp_mutans.rRNA 17702 114 + 2030929 --ACGAUGAGUGCUAGGUGUUAGGCCCUUUCCGGGGCUUAGUGCCGGAGCUAACGCAAUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGU--UGAAACUCAAAGGAAUUGACGGG--- --.(((((((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))--))..))))..))).)))).....--- ( -52.50) >sp_pneumo.rRNA 15973 114 + 2038623 --ACGAUGAGUGCUAGGUGUUAGACCCUUUCCGGGGUUUAGUGCCGUAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGU--UGAAACUCAAAGGAAUUGACGGG--- --.(((((((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))--))..))))..))).)))).....--- ( -50.50) >consensus __ACGAUGAGUGCUAGGUGUUAGGCCCUUUCCGGGGCUUAGUGCCGAAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGU__UGAAACUCAAAGGAAUUGACGGG___ ......((((((((.((..(((((((((....)))))))))..))...((....))....)))(((((......)))))...(((....)))......)))))................. (-23.51 = -25.95 + 2.44)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Fri Apr 7 15:50:29 2006