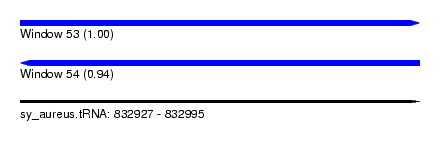
| Sequence ID | sy_aureus.tRNA |
|---|---|
| Location | 832,927 – 832,995 |
| Length | 68 |
| Max. P | 0.997459 |

| Location | 832,927 – 832,995 |
|---|---|
| Length | 68 |
| Sequences | 6 |
| Columns | 68 |
| Reading direction | forward |
| Mean pairwise identity | 91.08 |
| Mean single sequence MFE | -22.65 |
| Consensus MFE | -23.19 |
| Energy contribution | -22.22 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.53 |
| Structure conservation index | 1.02 |
| SVM decision value | 2.86 |
| SVM RNA-class probability | 0.997459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
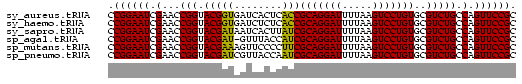
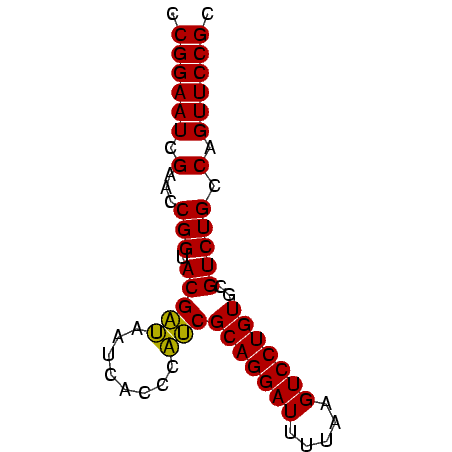
>sy_aureus.tRNA 832927 68 + 2809430 GCGGAACUGGCAGACGCACAGGACUUAAAAUCCUGCGGUGAGUGAUCACCGUACCGGUUCGAUUCCGG ...(((((((........(((((.......)))))((((((....))))))..)))))))........ ( -25.50) >sy_haemo.tRNA 184195 68 + 2685023 GCGGAACUGGCAGACGCACAGGACUUAAAAUCCUGCGGUGAGAGAUCACCGUACCGGUUCGAUUCCGG ...(((((((........(((((.......)))))((((((....))))))..)))))))........ ( -24.00) >sy_sapro.tRNA 204581 68 + 2516583 GCGGAACUGGCAGACGCACAGGACUUAAAAUCCUGCGAUAAGUGAUUAUCGUACCGGUUCGAUUCCGG ...(((((((........(((((.......)))))((((((....))))))..)))))))........ ( -21.20) >sp_agal.tRNA 21853 67 + 2160275 GCGGAACUGGCAGACGCACAGGACUUAAAAUCCUGCGAUGGUAAAC-AUCGUACCGGUUCGAUUCCGG ...(((((((........(((((.......)))))(((((.....)-))))..)))))))........ ( -20.90) >sp_mutans.tRNA 22565 68 + 2030929 GCGGAACUGGCAGACGCACAGGACUUAAAAUCCUGCGAAGGGGAACUUUCGUACCGGUUCGAUUCCGG ...(((((((........(((((.......)))))((((((....))))))..)))))))........ ( -22.80) >sp_pneumo.tRNA 249315 68 + 2038623 GCGGAACUGGCAGACGCACAGGACUUAAAAUCCUGCGAUUGGUAACGAUCGUACCGGUUCGAUUCCGG ...(((((((........(((((.......)))))((((((....))))))..)))))))........ ( -21.50) >consensus GCGGAACUGGCAGACGCACAGGACUUAAAAUCCUGCGAUGAGUAACCAUCGUACCGGUUCGAUUCCGG ...(((((((........(((((.......)))))((((((....))))))..)))))))........ (-23.19 = -22.22 + -0.97)

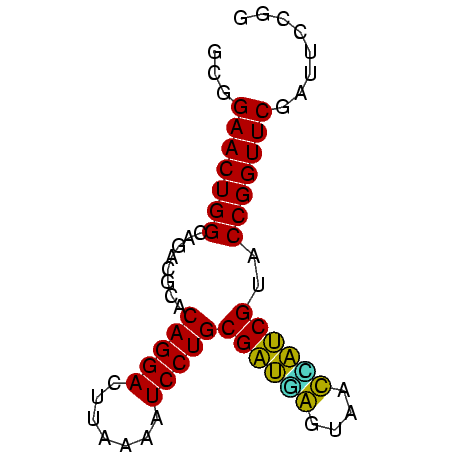
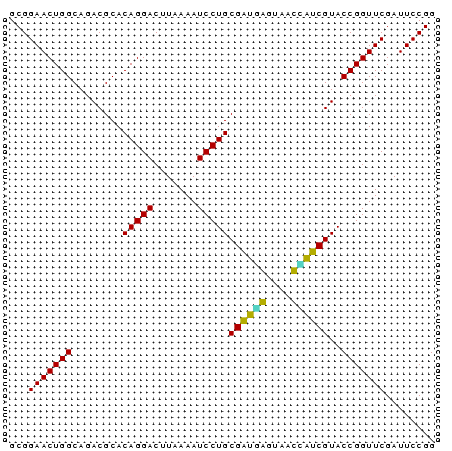
| Location | 832,927 – 832,995 |
|---|---|
| Length | 68 |
| Sequences | 6 |
| Columns | 68 |
| Reading direction | reverse |
| Mean pairwise identity | 91.08 |
| Mean single sequence MFE | -22.83 |
| Consensus MFE | -20.15 |
| Energy contribution | -19.43 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940904 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.tRNA 832927 68 - 2809430 CCGGAAUCGAACCGGUACGGUGAUCACUCACCGCAGGAUUUUAAGUCCUGUGCGUCUGCCAGUUCCGC .((((((.(...(((.(((((((....)))))(((((((.....)))))))..))))).).)))))). ( -25.00) >sy_haemo.tRNA 184195 68 - 2685023 CCGGAAUCGAACCGGUACGGUGAUCUCUCACCGCAGGAUUUUAAGUCCUGUGCGUCUGCCAGUUCCGC .((((((.(...(((.(((((((....)))))(((((((.....)))))))..))))).).)))))). ( -25.00) >sy_sapro.tRNA 204581 68 - 2516583 CCGGAAUCGAACCGGUACGAUAAUCACUUAUCGCAGGAUUUUAAGUCCUGUGCGUCUGCCAGUUCCGC .((((((.(...(((.(((((((....)))))(((((((.....)))))))..))))).).)))))). ( -20.70) >sp_agal.tRNA 21853 67 - 2160275 CCGGAAUCGAACCGGUACGAU-GUUUACCAUCGCAGGAUUUUAAGUCCUGUGCGUCUGCCAGUUCCGC ........((((.((((.(((-((........(((((((.....)))))))))))))))).))))... ( -22.10) >sp_mutans.tRNA 22565 68 - 2030929 CCGGAAUCGAACCGGUACGAAAGUUCCCCUUCGCAGGAUUUUAAGUCCUGUGCGUCUGCCAGUUCCGC .((((((.(((..((.((....)).))..)))(((((((.....)))))))..........)))))). ( -23.10) >sp_pneumo.tRNA 249315 68 - 2038623 CCGGAAUCGAACCGGUACGAUCGUUACCAAUCGCAGGAUUUUAAGUCCUGUGCGUCUGCCAGUUCCGC .((((((.(....(((((....).))))...((((((((.....)))))))).......).)))))). ( -21.10) >consensus CCGGAAUCGAACCGGUACGAUAAUCACCCAUCGCAGGAUUUUAAGUCCUGUGCGUCUGCCAGUUCCGC .((((((.(...(((.(((((........)))(((((((.....)))))))..))))).).)))))). (-20.15 = -19.43 + -0.72) # Strand winner: forward (0.89)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Fri Apr 7 15:51:13 2006