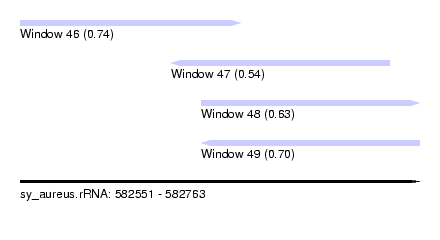
| Sequence ID | sy_aureus.rRNA |
|---|---|
| Location | 582,551 – 582,763 |
| Length | 212 |
| Max. P | 0.743276 |

| Location | 582,551 – 582,668 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.47 |
| Mean single sequence MFE | -38.10 |
| Consensus MFE | -37.51 |
| Energy contribution | -35.70 |
| Covariance contribution | -1.81 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.743276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 582551 117 + 2809430 CAUACCUCUGGUGUACCAGUUGUCGUGCCAACGGCAUAGCUGGGUAGCUAUGUGUGGACGGGAUAAGUGCUGAAAGCAUCUAAGCAUGAAGCCCCCCUCAAGAUGAGAUUUCCC---AAC (((((...((((...(((((((((((....))))))..)))))...)))).)))))...((((...(((((...........))))).........(((.....)))...))))---... ( -35.50) >sp_agal.rRNA 20916 120 + 2160275 CUUACCGCUGGUGUACCAGUUGUCUUGCCAAAGGCAUCGCUGGGUAGCUAUGUAGGGAAGGGAUAAGCGCUGAAAGCAUCUAAGUGCGAAGCCCACCUCAAGAUGAGAUUUCCCAUGAUU ......((((((...(((((((((((....))))))..)))))...)))).)).((((((((....(((((...........)))))....)))..(((.....)))..)))))...... ( -39.90) >sp_mutans.rRNA 21495 120 + 2030929 CUUACCGCUGGUGUACCAGUUGUUCUGCCAAGAGCAUCGCUGGGUAGCUAAGUAGGGAGGGGAUAAACGCUGAAAGCAUCUAAGUGUGAAGCCCCCCUCAAGAUGAGAUUUCCCAUAACG ......((((((...(((((((((((....))))))..)))))...))).))).((((((((....(((((...........)))))....)))).(((.....)))...))))...... ( -40.10) >sp_pneumo.rRNA 19626 120 + 2038623 CUUACCGCUGGUGUACCAGUUGUCUUGCCAAAGGCAUCGCUGGGUAGCUAUGUAGGGAAGGGAUAAACGCUGAAAGCAUCUAAGUGUGAAACCCACCUCAAGAUGAGAUUUCCCAUGAUU ......((((((...(((((((((((....))))))..)))))...)))).)).((((((((....(((((...........)))))....)))..(((.....)))..)))))...... ( -36.90) >consensus CUUACCGCUGGUGUACCAGUUGUCUUGCCAAAGGCAUCGCUGGGUAGCUAUGUAGGGAAGGGAUAAACGCUGAAAGCAUCUAAGUGUGAAGCCCACCUCAAGAUGAGAUUUCCCAUGAUU ..(((...((((...(((((((((((....))))))..)))))...)))).)))((((((((....(((((...........)))))....)))..(((.....)))..)))))...... (-37.51 = -35.70 + -1.81)

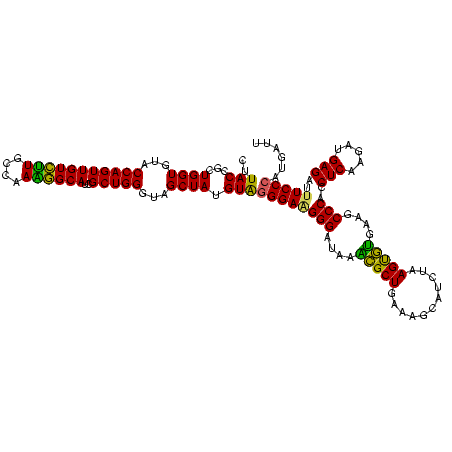
| Location | 582,631 – 582,747 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.42 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -17.48 |
| Energy contribution | -16.67 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.541337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 582631 116 - 2809430 AUUCGUCAGCUCCACAUGUCACCAUGCUUCCACCUCGAACCUAUUAACCUCAUCAUCUUUGAGGGAUCUUA-UAACCGAAGUU---GGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUA .......((((((.((((....))))......((((...........(((((.......))))).......-......(((.(---(((....)))).)))))))))))))......... ( -29.40) >sp_agal.rRNA 20996 120 - 2160275 AUUAGUCCGCUAAAUGUGUCACCACAAUUACACUCCUAACCUAUCUACCUGAUCAUCUCUCAGGGCUCUUACUGAUAUAAAAUCAUGGGAAAUCUCAUCUUGAGGUGGGCUUCGCACUUA ...(((((((....((((....)))).......(((((.........(((((.......)))))........((((.....)))))))))...(((.....))))))))))......... ( -25.80) >sp_mutans.rRNA 21575 120 - 2030929 AUUGGUCCGCUUCACGUCUCACAACGCUUCCACUCCCAACCUAUCUACCUGUUCUUCUCUCAGGGCUCUUACUAACUGAACGUUAUGGGAAAUCUCAUCUUGAGGGGGGCUUCACACUUA ...(((((.((((((((......))).......(((((.........((((.........))))........((((.....)))))))))..........))))).)))))......... ( -26.30) >sp_pneumo.rRNA 19706 120 - 2038623 AUUAGUCCGCUACAUGUGUCGCCACACUUCCACUUCUAACCUAUCUACCUGAUCAUCUCUCAGGGCUCUUACUGAUAUAUAAUCAUGGGAAAUCUCAUCUUGAGGUGGGUUUCACACUUA ..(((....)))..((((..(((.((((((.................(((((.......)))))..(((((.((((.....)))))))))...........)))))))))..)))).... ( -27.00) >consensus AUUAGUCCGCUACACGUGUCACCACACUUCCACUCCUAACCUAUCUACCUGAUCAUCUCUCAGGGCUCUUACUAACAGAAAAUCAUGGGAAAUCUCAUCUUGAGGGGGGCUUCACACUUA .((((.........((((....)))).........))))(((((((.(((((.......)))))......................(((....)))......)))))))........... (-17.48 = -16.67 + -0.81) # Strand winner: forward (0.99)

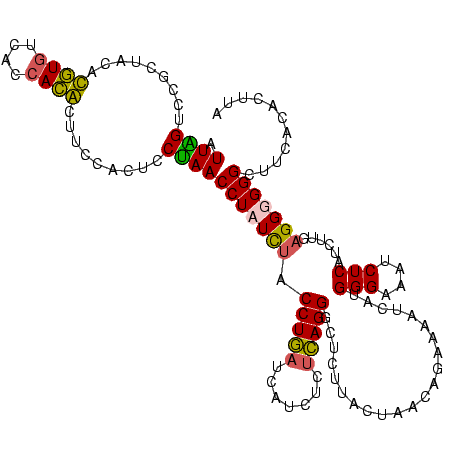
| Location | 582,647 – 582,763 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.25 |
| Mean single sequence MFE | -31.72 |
| Consensus MFE | -26.79 |
| Energy contribution | -26.23 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
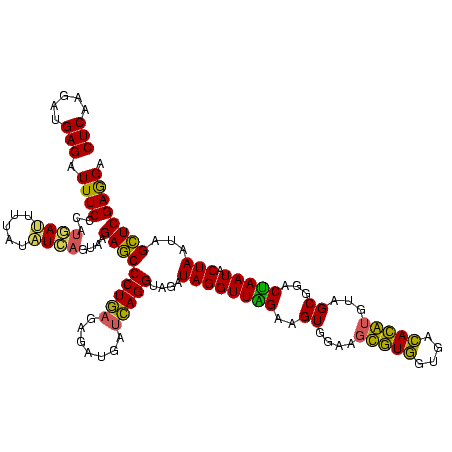
>sy_aureus.rRNA 582647 116 + 2809430 CUCAAGAUGAGAUUUCCC---AACUUCGGUUA-UAAGAUCCCUCAAAGAUGAUGAGGUUAAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUCGAUCGAAGA (((.....))).......---..((((((((.-.......(((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...)))))))). ( -33.80) >sp_agal.rRNA 21012 120 + 2160275 CUCAAGAUGAGAUUUCCCAUGAUUUUAUAUCAGUAAGAGCCCUGAGAGAUGAUCAGGUAGAUAGGUUAGGAGUGUAAUUGUGGUGACACAUUUAGCGGACUAAUACUAAUAGCUCGAGGA (((...((((((((......))))))))........(((((((((.......)))))....((((((((..((.(((.((((....)))).)))))...))))).)))...))))))).. ( -32.90) >sp_mutans.rRNA 21591 120 + 2030929 CUCAAGAUGAGAUUUCCCAUAACGUUCAGUUAGUAAGAGCCCUGAGAGAAGAACAGGUAGAUAGGUUGGGAGUGGAAGCGUUGUGAGACGUGAAGCGGACCAAUACUAAUCGCUCGAGGA (((.....)))...((((.((((.....))))....((((((((.........))))..((((((((((..((....(((((....)))))...))...))))).)).)))))))).))) ( -29.60) >sp_pneumo.rRNA 19722 120 + 2038623 CUCAAGAUGAGAUUUCCCAUGAUUAUAUAUCAGUAAGAGCCCUGAGAGAUGAUCAGGUAGAUAGGUUAGAAGUGGAAGUGUGGCGACACAUGUAGCGGACUAAUACUAAUAGCUCGAGGA (((.....)))...((((.((((.....))))....(((((((((.......)))))....((((((((..((....(((((....)))))...))...))))).)))...))))).))) ( -30.60) >consensus CUCAAGAUGAGAUUUCCCAUGAUUUUAUAUCAGUAAGAGCCCUGAGAGAUGAUCAGGUAGAUAGGUUAGAAGUGGAAGCGUGGUGACACAUGUAGCGGACUAAUACUAAUAGCUCGAGGA (((.....))).((((...((((.....))))....(((((((((.......)))))....((((((((..((....(((((....)))))...))...))))).)))...)))))))). (-26.79 = -26.23 + -0.56)


| Location | 582,647 – 582,763 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.25 |
| Mean single sequence MFE | -24.24 |
| Consensus MFE | -20.62 |
| Energy contribution | -19.74 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
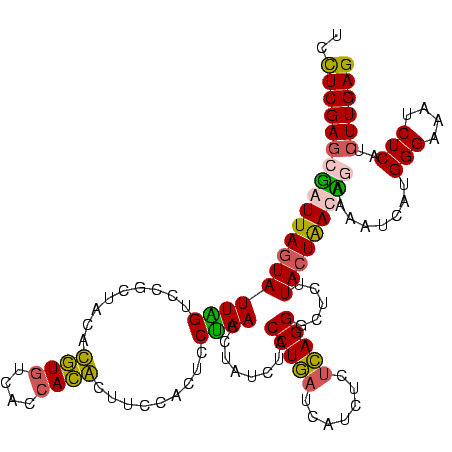
>sy_aureus.rRNA 582647 116 - 2809430 UCUUCGAUCGAUUAGUAUUCGUCAGCUCCACAUGUCACCAUGCUUCCACCUCGAACCUAUUAACCUCAUCAUCUUUGAGGGAUCUUA-UAACCGAAGUU---GGGAAAUCUCAUCUUGAG .....((((..((((((((((.........((((....)))).........))))..))))))(((((.......)))))))))...-......(((.(---(((....)))).)))... ( -22.17) >sp_agal.rRNA 21012 120 - 2160275 UCCUCGAGCUAUUAGUAUUAGUCCGCUAAAUGUGUCACCACAAUUACACUCCUAACCUAUCUACCUGAUCAUCUCUCAGGGCUCUUACUGAUAUAAAAUCAUGGGAAAUCUCAUCUUGAG ..((((((.((((((((..((((((.(((.((((....)))).))))...........(((.....))).........)))))..)))))))).......(((((....))))))))))) ( -26.90) >sp_mutans.rRNA 21591 120 - 2030929 UCCUCGAGCGAUUAGUAUUGGUCCGCUUCACGUCUCACAACGCUUCCACUCCCAACCUAUCUACCUGUUCUUCUCUCAGGGCUCUUACUAACUGAACGUUAUGGGAAAUCUCAUCUUGAG ..((((((.((((((..(((....((.....))....)))..)).....(((((.........((((.........))))........((((.....)))))))))))))....)))))) ( -21.50) >sp_pneumo.rRNA 19722 120 - 2038623 UCCUCGAGCUAUUAGUAUUAGUCCGCUACAUGUGUCGCCACACUUCCACUUCUAACCUAUCUACCUGAUCAUCUCUCAGGGCUCUUACUGAUAUAUAAUCAUGGGAAAUCUCAUCUUGAG ..((((((.((((((((..(((((.......((((....))))...............(((.....))).........)))))..)))))))).......(((((....))))))))))) ( -26.40) >consensus UCCUCGAGCGAUUAGUAUUAGUCCGCUACACGUGUCACCACACUUCCACUCCUAACCUAUCUACCUGAUCAUCUCUCAGGGCUCUUACUAACAGAAAAUCAUGGGAAAUCUCAUCUUGAG ..(((((((((((((((((((.........((((....)))).........))))........(((((.......))))).....)))))))))........(((....)))..)))))) (-20.62 = -19.74 + -0.88) # Strand winner: forward (1.00)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Fri Apr 7 15:51:09 2006