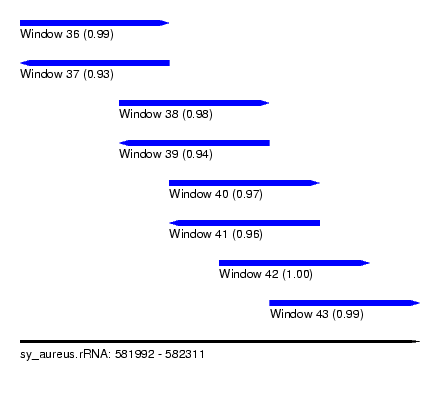
| Sequence ID | sy_aureus.rRNA |
|---|---|
| Location | 581,992 – 582,311 |
| Length | 319 |
| Max. P | 0.997744 |

| Location | 581,992 – 582,111 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.71 |
| Mean single sequence MFE | -38.20 |
| Consensus MFE | -38.49 |
| Energy contribution | -32.55 |
| Covariance contribution | -5.94 |
| Combinations/Pair | 1.65 |
| Mean z-score | -0.82 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 581992 119 + 2809430 AUAGGUAGGAGCCUUUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCU-UUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGG ((((.(((..(((((....((((((((....)))))))).)))))...(((((....)))))))).)))).(((((.-..((..(((.((((((.....)))))))).)..)).))))). ( -47.80) >sp_agal.rRNA 20357 119 + 2160275 AUAGGUAGGAGCCUAUGAUAACGGGACGCCAGUUUCGUUUGAGGCGUUGUUGGGAUACUACCCUUG-UGUUAUGGCUACUCUAACCCAGAUAGGCAAUCCCUAUCGGAGACAGUGUCUGA ((((((....)))))).......((((((..((((((((.(((..((..(...(((((.......)-)))))..))..))).)))...((((((.....)))))).))))).)))))).. ( -35.30) >sp_mutans.rRNA 20936 119 + 2030929 AUAGGUAGGAGCCAAGGAAGAGUGAACGCUAGUUUACUUGGAGGCGUUGUUGGGAUACUACCCUUG-UGUGAUGGCUACUCUAACCCGGUAGGUUGAUCAUCUACGGAGACAGUGUCUGA ...(((.((((........((((((((....))))))))..((.(((..(.(((......)))...-.)..))).)).)))).)))(.((((((.....)))))).)((((...)))).. ( -33.00) >sp_pneumo.rRNA 19067 119 + 2038623 AUAGGUAGGAGUCUAAGAGAUCGGGACGCCAGUUUCGAAGGAGACGCUGUUGGGAUACUACCCUUG-UGUUAUGGCCACUCUAACCCAGAUAGGUGAUCCCUAUCGGAGACAGUGUCUGA ...((((((..((..((((.(((..((....))..))).((.(((((....(((......)))..)-))))....)).)))).(((......)))))..))))))(....)......... ( -36.70) >consensus AUAGGUAGGAGCCUAUGAAAACGGAACGCCAGUUUACUUGGAGGCGCUGUUGGGAUACUACCCUUG_UGUGAUGGCUACUCUAACCCAGAUAGGUGAUCCCUAUCGGAGACAGUGUCUGA ...(((.(((((((.....((((((((....))))))))..))))((..(.(((......)))........)..))...))).)))..((((((.....))))))(....)......... (-38.49 = -32.55 + -5.94)



| Location | 581,992 – 582,111 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.71 |
| Mean single sequence MFE | -30.41 |
| Consensus MFE | -28.91 |
| Energy contribution | -25.48 |
| Covariance contribution | -3.44 |
| Combinations/Pair | 1.53 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933989 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 581992 119 - 2809430 CCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAA-AGCCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAGCGCUCACGUUUCAAAGGCUCCUACCUAU .(((((.(((.....((((((.....))))))))))))))..-((((......(((.(((......)))...)))......((((.(((....))).))))......))))......... ( -34.50) >sp_agal.rRNA 20357 119 - 2160275 UCAGACACUGUCUCCGAUAGGGAUUGCCUAUCUGGGUUAGAGUAGCCAUAACA-CAAGGGUAGUAUCCCAACAACGCCUCAAACGAAACUGGCGUCCCGUUAUCAUAGGCUCCUACCUAU ..((((...))))..((((((.....))))))(((((..((((....(((((.-...(((......)))....(((((............)))))...))))).....))))..))))). ( -33.90) >sp_mutans.rRNA 20936 119 - 2030929 UCAGACACUGUCUCCGUAGAUGAUCAACCUACCGGGUUAGAGUAGCCAUCACA-CAAGGGUAGUAUCCCAACAACGCCUCCAAGUAAACUAGCGUUCACUCUUCCUUGGCUCCUACCUAU .......(((.(((.((((.........)))).))).)))((.(((((.....-...(((......)))...(((((..............)))))..........))))).))...... ( -23.24) >sp_pneumo.rRNA 19067 119 - 2038623 UCAGACACUGUCUCCGAUAGGGAUCACCUAUCUGGGUUAGAGUGGCCAUAACA-CAAGGGUAGUAUCCCAACAGCGUCUCCUUCGAAACUGGCGUCCCGAUCUCUUAGACUCCUACCUAU ..((((.(((((((.((((((.....)))))).))).....(((.......))-)..(((......))).)))).))))...........((.(((...........))).))....... ( -30.00) >consensus UCAGACACUGUCUCCGAUAAGGAUCACCUAUCCGGGUUAGAGUAGCCAUAACA_CAAGGGUAGUAUCCCAACAACGCCUCCAACGAAACUAGCGUCCACUUUUCAUAGGCUCCUACCUAU .......(((.(((.((((((.....)))))).))).))).((((............(((......)))......((((..((((.(((....))).)))).....))))..)))).... (-28.91 = -25.48 + -3.44) # Strand winner: forward (1.00)

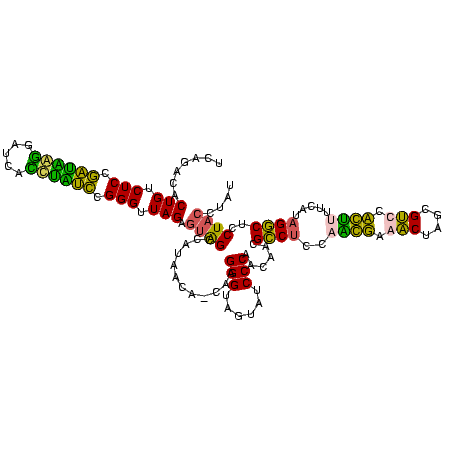
| Location | 582,071 – 582,191 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.33 |
| Mean single sequence MFE | -45.88 |
| Consensus MFE | -46.59 |
| Energy contribution | -43.90 |
| Covariance contribution | -2.69 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.91 |
| Structure conservation index | 1.02 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 582071 120 + 2809430 CUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGGCGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAU ((((((..((((((.....)))))).(((.....(((((((.....))))))).(((((((.(((((((...........))))))))).....)))))))).....))))))....... ( -44.60) >sp_agal.rRNA 20436 120 + 2160275 CUAACCCAGAUAGGCAAUCCCUAUCGGAGACAGUGUCUGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGGAAAUCAG ((((((..((((((.....))))))(....)...((((((.((((((.....)))((((....((((((...........)))))))))).......))))))))).))))))....... ( -48.00) >sp_mutans.rRNA 21015 120 + 2030929 CUAACCCGGUAGGUUGAUCAUCUACGGAGACAGUGUCUGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGGAAAUCAG ((((((..((((((.....))))))(....)...((((((.((((((.....)))((((....((((((...........)))))))))).......))))))))).))))))....... ( -45.50) >sp_pneumo.rRNA 19146 120 + 2038623 CUAACCCAGAUAGGUGAUCCCUAUCGGAGACAGUGUCUGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAU ((((((..((((((.....))))))(....)....(((((.((((((.....)))((((....((((((...........)))))))))).......))))))))..))))))....... ( -45.40) >consensus CUAACCCAGAUAGGUGAUCCCUAUCGGAGACAGUGUCUGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAG ((((((..((((((.....))))))(....)...((((((.((((((.....)))((((....((((((...........)))))))))).......))))))))).))))))....... (-46.59 = -43.90 + -2.69)

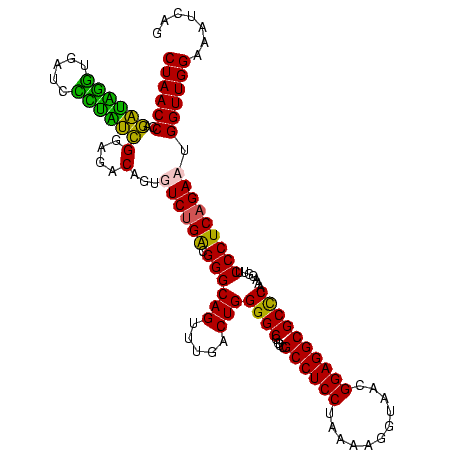

| Location | 582,071 – 582,191 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.33 |
| Mean single sequence MFE | -41.15 |
| Consensus MFE | -40.30 |
| Energy contribution | -38.92 |
| Covariance contribution | -1.37 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.940348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 582071 120 - 2809430 AUGAUUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAG ................(((.(((......((..(((((((...........)))))))..)))))))).......((((((..(((...)))....(((((.....)))))))))))... ( -38.10) >sp_agal.rRNA 20436 120 - 2160275 CUGAUUUCCAACCAGUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCGAUAGGGAUUGCCUAUCUGGGUUAG ..............(((((((((.......((((((((((...........))))))....))))(((.....)))))).)))))).(((.(((.((((((.....)))))).))).))) ( -44.90) >sp_mutans.rRNA 21015 120 - 2030929 CUGAUUUCCAACCAGUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCGUAGAUGAUCAACCUACCGGGUUAG ..............(((((((((.......((((((((((...........))))))....))))(((.....)))))).)))))).(((.(((.((((.........)))).))).))) ( -39.60) >sp_pneumo.rRNA 19146 120 - 2038623 AUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCGAUAGGGAUCACCUAUCUGGGUUAG ...............((((((((.......((((((((((...........))))))....))))(((.....)))))).)))))..(((.(((.((((((.....)))))).))).))) ( -42.00) >consensus AUGAUUUCCAACCAGUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCGAUAAGGAUCACCUAUCCGGGUUAG ..............(((((((((.......((.(((((((...........))))))).))....(((.....)))))).)))))).(((.(((.((((((.....)))))).))).))) (-40.30 = -38.92 + -1.37) # Strand winner: forward (1.00)

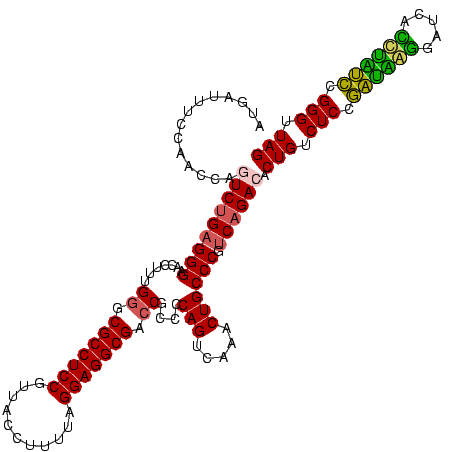

| Location | 582,111 – 582,231 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.69 |
| Mean single sequence MFE | -44.50 |
| Consensus MFE | -44.00 |
| Energy contribution | -43.12 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 582111 120 + 2809430 CGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGAC ....(((.....)))..((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((((....)))).)))))))))))))))..... ( -43.80) >sp_agal.rRNA 20476 120 + 2160275 CGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGGAAAUCAGUCGUAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAG ...((((((......((((....((((((...........))))))))))..(((((((((..(((((((.....)))))))......((((....)))))))))))))))))))..... ( -45.30) >sp_mutans.rRNA 21055 120 + 2030929 CGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGGAAAUCAGUCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAC ...((((((......((((....((((((...........))))))))))..(((((((((..(((((((.....)))))))((.....)).........)))))))))))))))..... ( -45.60) >sp_pneumo.rRNA 19186 120 + 2038623 CGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAG ...((((((......((((....((((((...........))))))))))..(((((((((..(((((((.....)))))))((.....)).........)))))))))))))))..... ( -43.30) >consensus CGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAGUCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAC ...((((((......((((....((((((...........))))))))))..(((((((((..(((((((.....))))))).....(((......))).)))))))))))))))..... (-44.00 = -43.12 + -0.88)


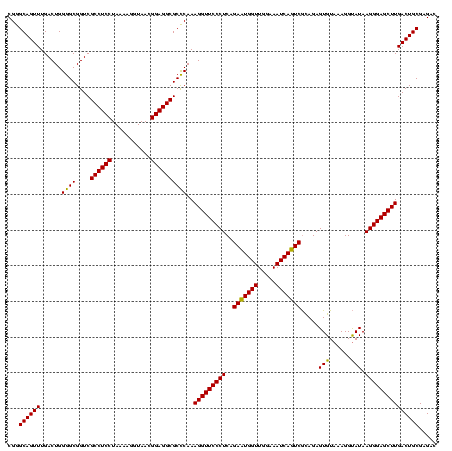
| Location | 582,111 – 582,231 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.69 |
| Mean single sequence MFE | -35.16 |
| Consensus MFE | -33.79 |
| Energy contribution | -33.54 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 582111 120 - 2809430 GUCUCGCAGUCAAGCUCCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCG .....(((((...((((.......((((((........(((((.........))))))))))).......))))((((((...........))))))(((.......)))..)))))... ( -32.34) >sp_agal.rRNA 20476 120 - 2160275 CUCUCGCAGUCAAGCUCCCUUAUACCUUUACACUCUACGACUGAUUUCCAACCAGUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCG .....(((((...(((((((((................(((((.........)))))))))))).....(((((((((((...........))))))).....))))))...)))))... ( -36.29) >sp_mutans.rRNA 21055 120 - 2030929 GUCUCGCAGUCAAGCUCCCUUAUACCUUUACACUCUGCGACUGAUUUCCAACCAGUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCG .....(((((...(((((((((........(.....).(((((.........)))))))))))).....(((((((((((...........))))))).....))))))...)))))... ( -36.40) >sp_pneumo.rRNA 19186 120 - 2038623 CUCUCGCAGUCAAGCUCCCUUAUACCUUUACACUCUGCGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCG (((((((((.........................))))(((((.........))))).))))).......((((((((((...........))))))(((.......))).....)))). ( -35.61) >consensus CUCUCGCAGUCAAGCUCCCUUAUACCUUUACACUCUACGAAUGAUUUCCAACCAGUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCG .....(((((...(((((((((................(((((.........))))))))))))......((.(((((((...........))))))).))......))...)))))... (-33.79 = -33.54 + -0.25) # Strand winner: forward (1.00)


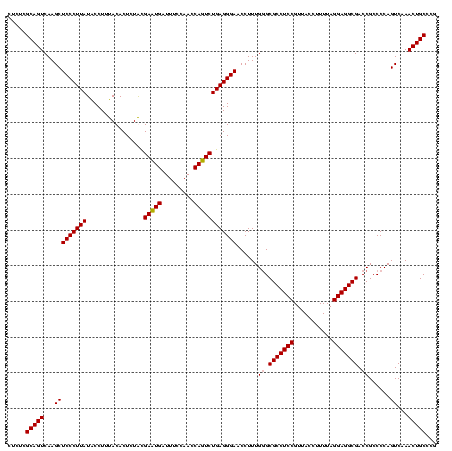
| Location | 582,151 – 582,271 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -44.47 |
| Consensus MFE | -43.88 |
| Energy contribution | -42.38 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.997744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 582151 120 + 2809430 GGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUGAUCC (((..((((...(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....)))......)))).))) ( -41.70) >sp_agal.rRNA 20516 120 + 2160275 GGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGGAAAUCAGUCGUAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCC (((.((((((..(((((((((..(((((((.....)))))))......((((....)))))))))))))..((((..(((......)))..)))).(((....)))))))...))..))) ( -45.80) >sp_mutans.rRNA 21095 120 + 2030929 GGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGGAAAUCAGUCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGACAGACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCC (((.((((((..(((((((((..(((((((.....)))))))((.....)).........)))))))))..((((..(((......)))..)))).(((....)))))))...))..))) ( -46.60) >sp_pneumo.rRNA 19226 120 + 2038623 GGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCC (((.((((((..(((((((((..(((((((.....)))))))((.....)).........)))))))))..((((..(((......)))..)))).(((....)))))))...))..))) ( -43.80) >consensus GGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAGUCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGACCUACAACUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCC (((.((((((..(((((((((..(((((((.....))))))).....(((......))).)))))))))..((((..(((......)))..)))).(((....)))))))...))..))) (-43.88 = -42.38 + -1.50)

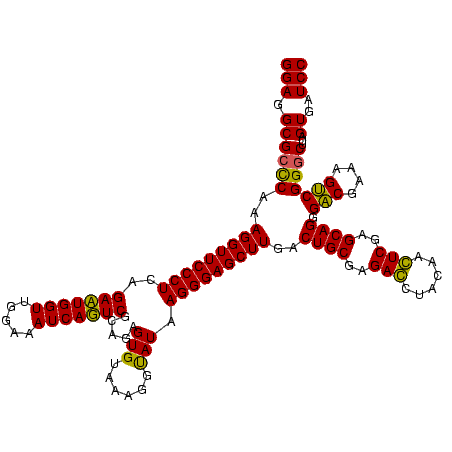
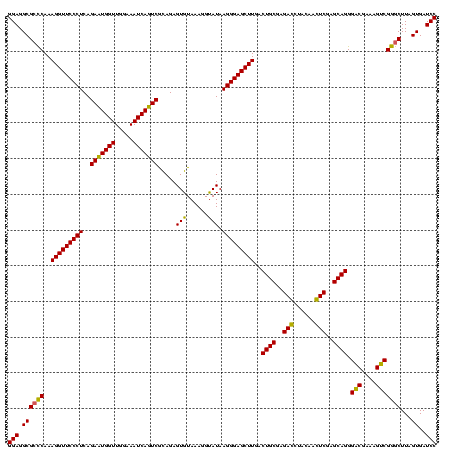
| Location | 582,191 – 582,311 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -41.27 |
| Consensus MFE | -39.88 |
| Energy contribution | -40.00 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_aureus.rRNA 582191 120 + 2809430 UCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAAAA .....(((((....(((.((((...((((((((............))))))))...(((....)))...))))....(((((((....))).))))...)))..)))))........... ( -38.50) >sp_agal.rRNA 20556 120 + 2160275 UCGUAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGUAUGGAAGGGCCAUCGCUCAACGGAUAAAA ((((.((((.....((.((....((((((..((((..(((......)))..)))).(((....)))))))))....))))((((((.((........)))))))))))).))))...... ( -43.50) >sp_mutans.rRNA 21135 120 + 2030929 UCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGACAGACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUACCGUAUGGAAGGGCCAUCGCUCAACGGAUAAAA (((..((((.....((.((....((((((..((((..(((......)))..)))).(((....)))))))))....))))((((((.((........))))))))))))..)))...... ( -41.80) >sp_pneumo.rRNA 19266 120 + 2038623 UCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGUAUGGAAGGGCCAUCGCUCAACGGAUAAAA (((..((((.....((.((....((((((..((((..(((......)))..)))).(((....)))))))))....))))((((((.((........))))))))))))..)))...... ( -41.30) >consensus UCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGACCUACAACUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGUAUGGAAGGGCCAUCGCUCAACGGAUAAAA ((((.((((.....((.((....((((((..((((..(((......)))..)))).(((....)))))))))....))))((((((.((........)))))))))))).))))...... (-39.88 = -40.00 + 0.12)

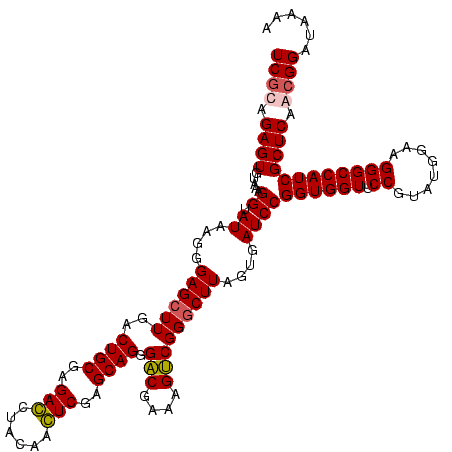

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Fri Apr 7 15:51:03 2006