
| Sequence ID | sy_ha.0 |
|---|---|
| Location | 184,579 – 184,699 |
| Length | 120 |
| Max. P | 0.999995 |

| Location | 184,579 – 184,674 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 59.04 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -7.60 |
| Energy contribution | -7.35 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.27 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
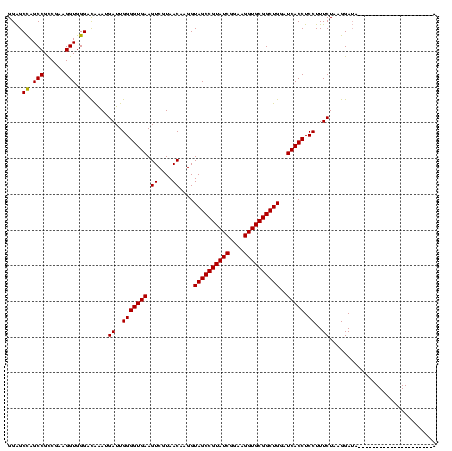
>sy_ha.0 184579 95 - 2685015/40-160 ------ACAGUAACUU-------------AAAUGUUACA---AACACUAUUUAGUAUU--AUGAGCUAAUCAAACAUCAUAAAUU-UUUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAAC ------...(((((..-------------....))))).---............((((--.((((((((((((((.((((((...-.))))))...)))))))..))))))).))))... ( -24.40) >sy_sa.0 204973 117 - 2516575/40-160 G---CAACAUUAAAUUGUUGGUUCAAAACAAGUGUUAGAGAUAACACAAAUUAGUAUUUUAUGAGCUAAUCAAACAUCAUAAUUCAUUUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAAC .---(((((......)))))(((((......((((((....))))))..............((((((((((((((.((((((.....))))))...)))))))..)))))))...))))) ( -32.70) >sy_au.0 572678 95 + 2809422/40-160 -----AAACGUAACU--------------AUAAGUUACA---AACAUUAUUUAGUAUUU-AUGAGCUAAUCAAACAUCAUAAUUU--UUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAAC -----....(((((.--------------....))))).---............(((((-.((((((((((((((.(((((....--.)))))...)))))))..))))))))))))... ( -22.10) >sp_ag.0 22124 111 + 2160267/40-160 G----GAUUUACAUUGUAAAUCCGU-CAAUGACAAAAAACAAUAAAUCUGUCAGCGAGACAGAAAAGAGCAA-AGCUCAAACUUU--UAA-UGAGAGUUUGAUCCUGGCUCAGGACGAAC (----(((((((...))))))))((-(...................((((((.....))))))...((((..-.(.(((((((((--(..-.)))))))))).)...))))..))).... ( -34.90) >sp_pn.0 341324 114 - 2038615/40-160 GCUACAACAUAAGUUGUAGUACUGAACAAUGA-AAAAAACAAUAAAUCUGUCAGUGA--CAGAAAUGAGUAAGAACUCAAACUUU--UAA-UGAGAGUUUGAUCCUGGCUCAGGACGAAC ((((((((....))))))))......((.(((-((...........((((((...))--))))..(((((....)))))...)))--)).-))...(((((.(((((...)))))))))) ( -30.30) >sp_mu.0 152460 111 - 2030921/40-160 G--ACAUAAAGAAAUGUGAC-CUGU-CAAAGA--ACGAACCAUAAAUCUGUCAGUGG-ACAGUAAUGAGAAAGAACUCAAACGAU--UAAAUGAGAGUUUGAUCCUGGCUCAGGACGAAC .--((((......))))..(-(((.-...((.--.............(((((....)-))))...((((......))))...(((--(((((....))))))))))....))))...... ( -23.50) >consensus G____AACAUUAAAUGU____CUG__CAAAGA_GAUAAACAAAAAAUCAGUCAGUAUU_CAGGAACGAAUAAAACAUCAAAAUUU__UAAUGGAGAGUUUGAUCCUGGCUCAGGACGAAC ................................................................................................(((((.(((((...)))))))))) ( -7.60 = -7.35 + -0.25)

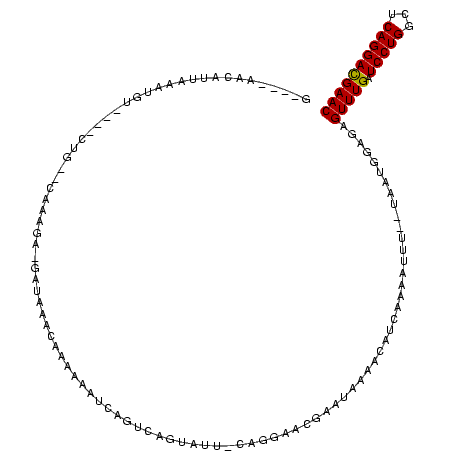
| Location | 184,579 – 184,674 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 59.04 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -6.30 |
| Energy contribution | -6.05 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.24 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.892516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 184579 95 - 2685015/40-160 GUUCAUCCUGAGCCAGGAUCAAACUCUCCAUAAA-AAUUUAUGAUGUUUGAUUAGCUCAU--AAUACUAAAUAGUGUU---UGUAACAUUU-------------AAGUUACUGU------ ........(((((...((((((((....(((((.-...)))))..)))))))).))))).--((((((....))))))---.(((((....-------------..)))))...------ ( -22.80) >sy_sa.0 204973 117 - 2516575/40-160 GUUCAUCCUGAGCCAGGAUCAAACUCUCCAUAAAUGAAUUAUGAUGUUUGAUUAGCUCAUAAAAUACUAAUUUGUGUUAUCUCUAACACUUGUUUUGAACCAACAAUUUAAUGUUG---C (((((...(((((...((((((((....(((((.....)))))..)))))))).))))).........(((..((((((....))))))..))).)))))(((((......)))))---. ( -28.80) >sy_au.0 572678 95 + 2809422/40-160 GUUCAUCCUGAGCCAGGAUCAAACUCUCCAUAA--AAAUUAUGAUGUUUGAUUAGCUCAU-AAAUACUAAAUAAUGUU---UGUAACUUAU--------------AGUUACGUUU----- ........(((((...((((((((....((((.--....))))..)))))))).))))).-.................---.(((((....--------------.)))))....----- ( -20.10) >sp_ag.0 22124 111 + 2160267/40-160 GUUCGUCCUGAGCCAGGAUCAAACUCUCA-UUA--AAAGUUUGAGCU-UUGCUCUUUUCUGUCUCGCUGACAGAUUUAUUGUUUUUUGUCAUUG-ACGGAUUUACAAUGUAAAUC----C ....(((..((((.(((.(((((((....-...--..))))))).))-).))))...((((((.....))))))...................)-))((((((((...)))))))----) ( -31.40) >sp_pn.0 341324 114 - 2038615/40-160 GUUCGUCCUGAGCCAGGAUCAAACUCUCA-UUA--AAAGUUUGAGUUCUUACUCAUUUCUG--UCACUGACAGAUUUAUUGUUUUUU-UCAUUGUUCAGUACUACAACUUAUGUUGUAGC .......((((((..(((..((((.....-...--......(((((....)))))..((((--((...))))))......))))..)-))...))))))..(((((((....))))))). ( -28.20) >sp_mu.0 152460 111 - 2030921/40-160 GUUCGUCCUGAGCCAGGAUCAAACUCUCAUUUA--AUCGUUUGAGUUCUUUCUCAUUACUGU-CCACUGACAGAUUUAUGGUUCGU--UCUUUG-ACAG-GUCACAUUUCUUUAUGU--C ....(.((((((((((((((((((.........--...)))))).)))..........((((-(....))))).....))))))((--(....)-))))-).)((((......))))--. ( -23.50) >consensus GUUCAUCCUGAGCCAGGAUCAAACUCUCAAUAA__AAAGUAUGAGGUUUGACUAGCUCAUG_AACACUAACAGAUGUAUUGUUUAAC_UCUUUG__CAG____ACAAUUAAUGUU____C (((.((((((...))))))..)))................................................................................................ ( -6.30 = -6.05 + -0.25) # Strand winner: forward (0.86)

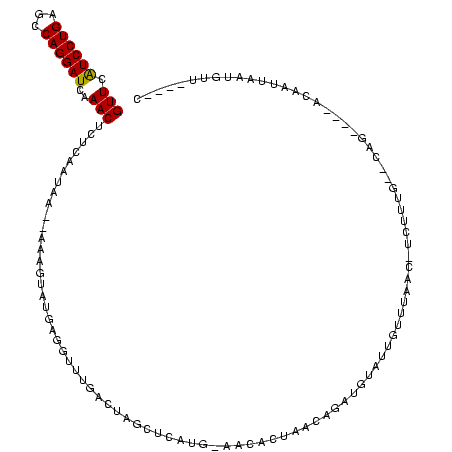
| Location | 184,579 – 184,694 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.60 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -16.19 |
| Energy contribution | -15.56 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 184579 115 - 2685015/80-200 --AACACUAUUUAGUAUU--AUGAGCUAAUCAAACAUCAUAAAUU-UUUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGAACAGA --............((((--.((((((((((((((.((((((...-.))))))...)))))))..))))))).))))..(((((((.(((((.......)))))..))).))))...... ( -32.90) >sy_sa.0 204973 120 - 2516575/80-200 AUAACACAAAUUAGUAUUUUAUGAGCUAAUCAAACAUCAUAAUUCAUUUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGAACAGA ..............((((...((((((((((((((.((((((.....))))))...)))))))..))))))).))))..(((((((.(((((.......)))))..))).))))...... ( -32.40) >sy_au.0 572678 115 + 2809422/80-200 --AACAUUAUUUAGUAUUU-AUGAGCUAAUCAAACAUCAUAAUUU--UUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGAACGGA --............(((((-.((((((((((((((.(((((....--.)))))...)))))))..))))))))))))..(((((((.(((((.......)))))..))).))))...... ( -31.00) >sp_ag.0 22124 116 + 2160267/80-200 AAUAAAUCUGUCAGCGAGACAGAAAAGAGCAA-AGCUCAAACUUU--UAA-UGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUAGAACGCUGAGG ..........((((((..........((((..-.(.(((((((((--(..-.)))))))))).)...))))(((....((((.....)))).))).................)))))).. ( -33.60) >sp_pn.0 341324 115 - 2038615/80-200 AAUAAAUCUGUCAGUGA--CAGAAAUGAGUAAGAACUCAAACUUU--UAA-UGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUAGAACGCUGAAG .........(((((((.--......(((((.((...(((((((((--(..-.))))))))))..)).))))).......)))))))(((((......(((......)))..))))).... ( -32.14) >sp_mu.0 152460 117 - 2030921/80-200 CAUAAAUCUGUCAGUGG-ACAGUAAUGAGAAAGAACUCAAACGAU--UAAAUGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUGGGACGCAAGGA ......((((((((((.-...((..((((..((...((((((..(--(....))..))))))..))..))))..))...))))))).((((.((..............)).))))..))) ( -35.24) >consensus AAAAAAUCAGUCAGUAUU_CAGGAACGAAUAAAACAUCAAAAUUU__UAAUGGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAACGAACAGA ..........(((((.....................((..............))..(((((.(((((...)))))))))))))))..(((((.......)))))................ (-16.19 = -15.56 + -0.64)

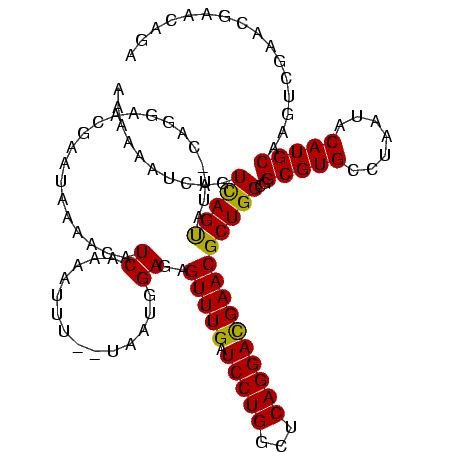
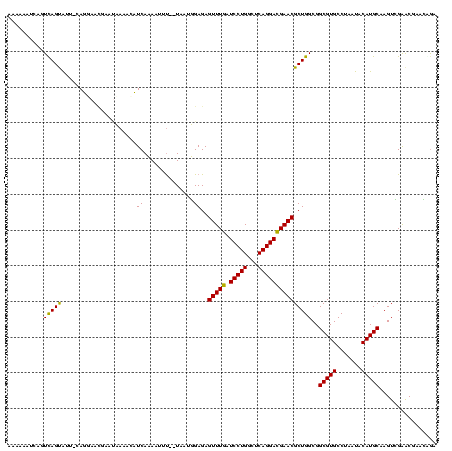
| Location | 184,579 – 184,691 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.52 |
| Mean single sequence MFE | -35.53 |
| Consensus MFE | -23.39 |
| Energy contribution | -19.75 |
| Covariance contribution | -3.64 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 184579 112 - 2685015/200-320 CAAG----GAGCUUGCUC----CUUUGACGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUU ...(----..(((..(((----.((((.((....)).)))).)))..)))..)..(((((((...)))))))....((((..((.((((((....)))))).))...))))......... ( -37.30) >sy_sa.0 204973 112 - 2516575/200-320 UAAG----GAGCUUGCUC----CUUUGACGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAACAUUU ...(----..(((..(((----.((((.((....)).)))).)))..)))..)..(((((((...)))))))....((((..((.((((((....)))))).))...))))......... ( -37.30) >sy_au.0 572678 112 + 2809422/200-320 CGAG----AAGCUUGCUU----CUCUGAUGUUAGCGGCGGACGGGUGAGUAACACGUGGAUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUU .(((----(((....)))----))).(((((((.(((.((.((.(((.....))).)).....))..((((......))))....((((((....))))))......)))..))))))). ( -33.30) >sp_ag.0 22124 112 + 2160267/200-320 UUUG----GUGUUUACAC----UAGACUGAUGAGUUGCGAACGGGUGAGUAACGCGUAGGUAACCUGCCUCAUAGCGGGGGAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAGAGUA ((((----(((....)))----)))).........(((..(((.((.....)).))).((((.(((((......)))))......((((((....))))))....)))))))........ ( -33.80) >sp_pn.0 341324 110 - 2038615/200-320 GA-G----GAGCUUGC-U----UCUCUGGAUGAGUUGCGAACGGGUGAGUAACGCGUAGGUAACCUGCCUGGUAGCGGGGGAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAGAGUG ((-(----(((....)-)----)))).........(((..(((.((.....)).))).((((.(((((......)))))......((((((....))))))....)))))))........ ( -35.60) >sp_mu.0 152460 120 - 2030921/200-320 AACACACUGUGCUUGCACACCGUGUUUUCUUGAGUCGCGAACGGGUGAGUAACGCGUAGGUAACCUGCCUAUUAGCGGGGGAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAUAUUA (((((..((((....))))..)))))..........((..(((.((.....)).))).((((.(((((......)))))......((((((....))))))....))))))......... ( -35.90) >consensus CAAG____GAGCUUGCUC____CUUUGACGUGAGCGGCGAACGGGUGAGUAACACGUAGGUAACCUACCUAUAAGACGGGGAUAACUACGGGAAACCAGAGCUAAUACCGCAUAAUAUUU ((((....(((....)))....))))..............(((.((.....)).)))(((((...)))))......(.((..((.((((((....)))))).))...)).)......... (-23.39 = -19.75 + -3.64)

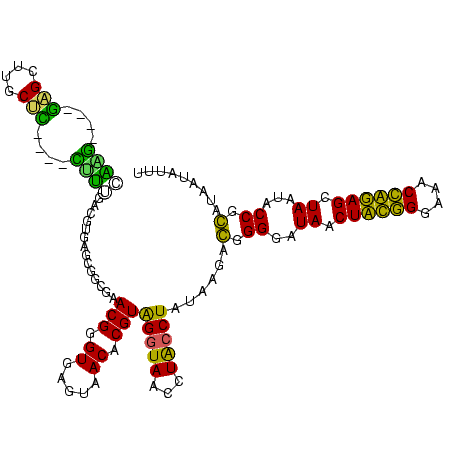
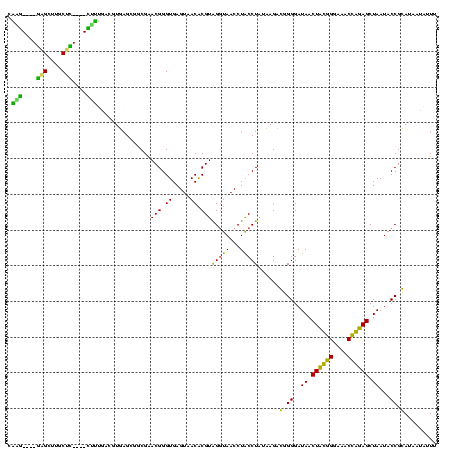
| Location | 184,579 – 184,699 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.30 |
| Mean single sequence MFE | -38.33 |
| Consensus MFE | -32.48 |
| Energy contribution | -26.52 |
| Covariance contribution | -5.97 |
| Combinations/Pair | 1.75 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 184579 120 - 2685015/240-360 ACGGGUGAGUAACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUCGAACCGCAUGGUUCGAUAGUGAAAGAUGGUUUUGCUAUC ............((((((((((...)))))))....((((..((.((((((....)))))).))...))))........(((((((....)))))))..)))...(((((.....))))) ( -38.30) >sy_sa.0 204973 120 - 2516575/240-360 ACGGGUGAGUAACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAACAUUUGGAACCGCAUGGUUCUAAAGUGAAAGAUGGUUUUGCUAUC ............((((((((((...)))))))....((((..((.((((((....)))))).))...))))......(((((((((....))))))))))))...(((((.....))))) ( -38.70) >sy_au.0 572678 120 + 2809422/240-360 ACGGGUGAGUAACACGUGGAUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUUGAACCGCAUGGUUCAAAAGUGAAAGACGGUCUUGCUGUC (((((((.....)))((((.....))))...((((((((((.((.((((((....)))))).))...))........(((((((((....)))))))))........)))))))))))). ( -37.10) >sp_ag.0 22124 119 + 2160267/240-360 ACGGGUGAGUAACGCGUAGGUAACCUGCCUCAUAGCGGGGGAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAGAGUAAUUAACACAUGUUAGUUAUUUAAAAGGAGCAAUUGC-UUC (((.((.....)).))).((((.(((((......)))))......((((((....))))))....))))......(((((((((((....)))))))))))....(((((....))-))) ( -38.00) >sp_pn.0 341324 119 - 2038615/240-360 ACGGGUGAGUAACGCGUAGGUAACCUGCCUGGUAGCGGGGGAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAGAGUGGAUGUUGCAUGACAUUUGCUUAAAAGGUGCACUUGC-AUC ....(..(((........((((.(((((......)))))......((((((....))))))....))))((((..(((..((((((....))))))..))).....)))))))..)-... ( -41.30) >sp_mu.0 152460 119 - 2030921/240-360 ACGGGUGAGUAACGCGUAGGUAACCUGCCUAUUAGCGGGGGAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAUAUUAAUUAUUGCAUGAUAAUUGAUUGAAAGAUGCAAGCGC-AUC .............((((.((((.(((((......)))))......((((((....))))))....))))((((...((((((((((....))))))))))......))))..))))-... ( -36.60) >consensus ACGGGUGAGUAACACGUAGGUAACCUACCUAUAAGACGGGGAUAACUACGGGAAACCAGAGCUAAUACCGCAUAAUAUUUAGAACCGCAUGGUACUAAAGUGAAAGAUGCAAUUGC_AUC ..((((..((.((......)).))..))))......(.((..((.((((((....)))))).))...)).)......(((((((((....)))))))))..................... (-32.48 = -26.52 + -5.97)
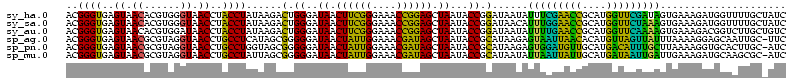
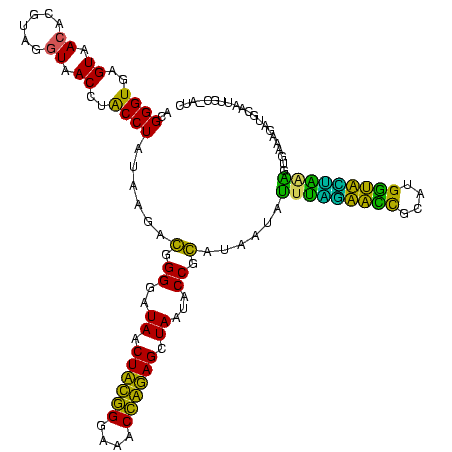
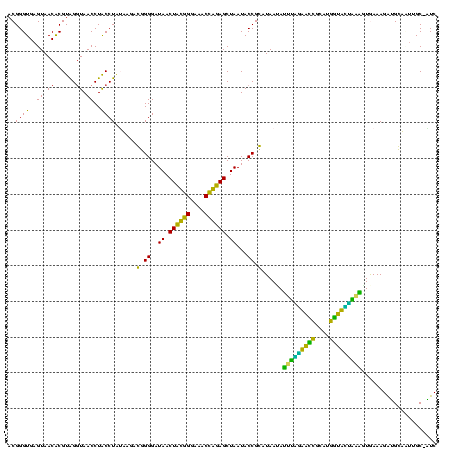
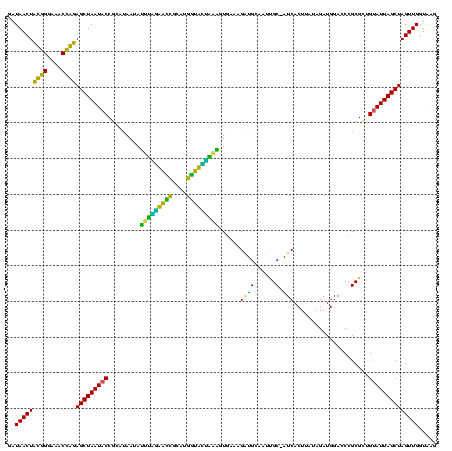
| Location | 184,579 – 184,699 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.57 |
| Mean single sequence MFE | -38.50 |
| Consensus MFE | -32.64 |
| Energy contribution | -26.70 |
| Covariance contribution | -5.94 |
| Combinations/Pair | 1.71 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 184579 120 - 2685015/280-400 GAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUCGAACCGCAUGGUUCGAUAGUGAAAGAUGGUUUUGCUAUCACUUAUAGAUGGACCCGCGCCGUAUUAGCUAGUUGGUAAG ..(((((((((....))))(((((((((((((......))))...(((..((((((((((..(((.....)))..))))).(.....)..))))).)))..))))))))))))))..... ( -39.90) >sy_sa.0 204973 120 - 2516575/280-400 GAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAACAUUUGGAACCGCAUGGUUCUAAAGUGAAAGAUGGUUUUGCUAUCACUUAUAGAUGGACCCGCGCCGUAUUAGCUAGUUGGUAAG ..(((((((((....))))(((((((((.((......(((((((((....)))))))))(((...(((((.....))))).((.......))...))).))))))))))))))))..... ( -39.20) >sy_au.0 572678 120 + 2809422/280-400 GAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUUGAACCGCAUGGUUCAAAAGUGAAAGACGGUCUUGCUGUCACUUAUAGAUGGAUCCGCGCUGCAUUAGCUAGUUGGUAAG .....((((((....))))))....((((((((....(((((((((....)))))))))((((..(((((.....)))))..))))......))))..(((((....)).))).)))).. ( -39.40) >sp_ag.0 22124 119 + 2160267/280-400 GAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAGAGUAAUUAACACAUGUUAGUUAUUUAAAAGGAGCAAUUGC-UUCACUGUGAGAUGGACCUGCGUUGUAUUAGCUAGUUGGUGAG ..(((((((((....))))(((((((((((((...(((((((((((....)))))))))))....(((((....))-)))...............))))..))))))))))))))..... ( -37.10) >sp_pn.0 341324 119 - 2038615/280-400 GAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAGAGUGGAUGUUGCAUGACAUUUGCUUAAAAGGUGCACUUGC-AUCACUACCAGAUGGACCUGCGUUGUAUUAGCUAGUUGGUGGG ..(((((((((....))))(((((((((((((...(((..((((((....))))))..)))....(((((....))-)))....((....))...))))..))))))))))))))..... ( -40.20) >sp_mu.0 152460 119 - 2030921/280-400 GAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAUAUUAAUUAUUGCAUGAUAAUUGAUUGAAAGAUGCAAGCGC-AUCACUAGUAGAUGGACCUGCGUUGUAUUAGCUAGUUGGUAAG ..(((((((((....))))(((((((((.((((...((((((((((....))))))))))......)))).(((((-(...(((.....)))...))))))))))))))))))))..... ( -35.20) >consensus GAUAACUACGGGAAACCAGAGCUAAUACCGCAUAAUAUUUAGAACCGCAUGGUACUAAAGUGAAAGAUGCAAUUGC_AUCACUUAUAGAUGGACCCGCGCUGUAUUAGCUAGUUGGUAAG ..(((((((((....))))(((((((((.........(((((((((....)))))))))......((((......).))).....................))))))))))))))..... (-32.64 = -26.70 + -5.94)

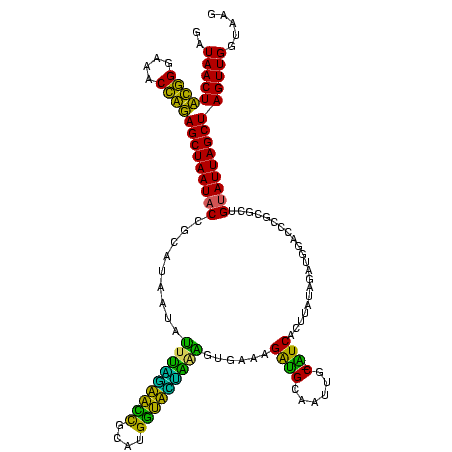
| Location | 184,579 – 184,699 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.57 |
| Mean single sequence MFE | -28.73 |
| Consensus MFE | -18.22 |
| Energy contribution | -16.70 |
| Covariance contribution | -1.52 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
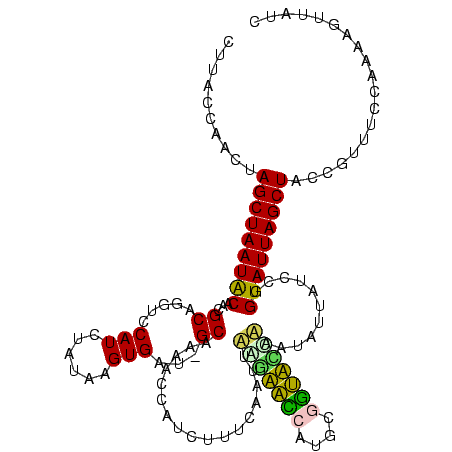
>sy_ha.0 184579 120 - 2685015/280-400 CUUACCAACUAGCUAAUACGGCGCGGGUCCAUCUAUAAGUGAUAGCAAAACCAUCUUUCACUAUCGAACCAUGCGGUUCGAAAUAUUAUCCGGUAUUAGCUCCGGUUUCCCGAAGUUAUC ......(((((((((((((....(((((.........(((((.((.........)).))))).(((((((....)))))))......)))))))))))))).(((....))).))))... ( -32.60) >sy_sa.0 204973 120 - 2516575/280-400 CUUACCAACUAGCUAAUACGGCGCGGGUCCAUCUAUAAGUGAUAGCAAAACCAUCUUUCACUUUAGAACCAUGCGGUUCCAAAUGUUAUCCGGUAUUAGCUCCGGUUUCCCGAAGUUAUC ......(((((((((((((.(.((((((...((((.((((((.((.........)).))))))))))))).)))(((..(....)..)))).))))))))).(((....))).))))... ( -29.70) >sy_au.0 572678 120 + 2809422/280-400 CUUACCAACUAGCUAAUGCAGCGCGGAUCCAUCUAUAAGUGACAGCAAGACCGUCUUUCACUUUUGAACCAUGCGGUUCAAAAUAUUAUCCGGUAUUAGCUCCGGUUUCCCGAAGUUAUC ......(((((((((((((....(((((..........((((.(((......).)).))))(((((((((....)))))))))....)))))))))))))).(((....))).))))... ( -33.00) >sp_ag.0 22124 119 + 2160267/280-400 CUCACCAACUAGCUAAUACAACGCAGGUCCAUCUCACAGUGAA-GCAAUUGCUCCUUUUAAAUAACUAACAUGUGUUAAUUACUCUUAUGCGGUAUUAGCUAUCGUUUCCAAUAGUUAUC .........((((((((((..((((((......))..((((((-((....))).........((((........)))).)))))....)))))))))))))).................. ( -20.80) >sp_pn.0 341324 119 - 2038615/280-400 CCCACCAACUAGCUAAUACAACGCAGGUCCAUCUGGUAGUGAU-GCAAGUGCACCUUUUAAGCAAAUGUCAUGCAACAUCCACUCUUAUGCGGUAUUAGCUAUCGUUUCCAAUAGUUAUC .........((((((((((..(((((((.(((...(((....)-))..))).)))......(((.......)))..............)))))))))))))).................. ( -26.70) >sp_mu.0 152460 119 - 2030921/280-400 CUUACCAACUAGCUAAUACAACGCAGGUCCAUCUACUAGUGAU-GCGCUUGCAUCUUUCAAUCAAUUAUCAUGCAAUAAUUAAUAUUAUGCGGUAUUAGCUAUCGUUUCCAAUAGUUAUC .........((((((((((...((((((.((((.......)))-).))))))...................((((.((((....)))))))))))))))))).................. ( -29.60) >consensus CUUACCAACUAGCUAAUACAACGCAGGUCCAUCUAUAAGUGAU_GCAAAACCAUCUUUCAAUUAAGAACCAUGCGGUACCAAAUAUUAUCCGGUAUUAGCUACCGUUUCCAAAAGUUAUC ..........(((((((((...((.....(((......)))...)).................(((((((....)))))))...........)))))))))................... (-18.22 = -16.70 + -1.52) # Strand winner: forward (1.00)


| Location | 184,579 – 184,699 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.28 |
| Mean single sequence MFE | -41.93 |
| Consensus MFE | -39.81 |
| Energy contribution | -40.37 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
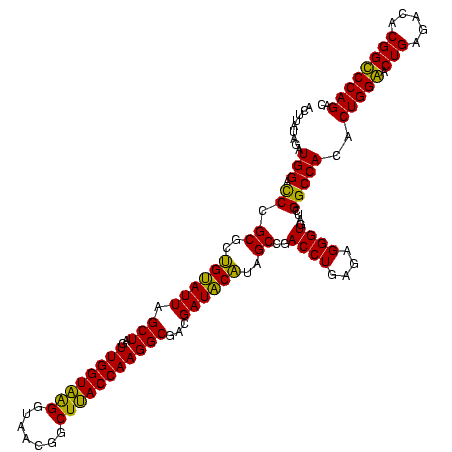
>sy_ha.0 184579 120 - 2685015/360-480 ACUUAUAGAUGGACCCGCGCCGUAUUAGCUAGUUGGUAAGGUAACGGCUUACCAAGGCGACGAUACGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGAACUGAGACACGGUCCAGAC .........((((((((((((((....))...((((((((.......)))))))))))).))....((.((((((((....)))...))))).))................))))))... ( -41.90) >sy_sa.0 204973 120 - 2516575/360-480 ACUUAUAGAUGGACCCGCGCCGUAUUAGCUAGUUGGUAAGGUAACGGCUUACCAAGGCGACGAUACGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGAACUGAGACACGGUCCAGAC .........((((((((((((((....))...((((((((.......)))))))))))).))....((.((((((((....)))...))))).))................))))))... ( -41.90) >sy_au.0 572678 120 + 2809422/360-480 ACUUAUAGAUGGAUCCGCGCUGCAUUAGCUAGUUGGUAAGGUAACGGCUUACCAAGGCAACGAUGCAUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGAACUGAGACACGGUCCAGAC .........((((.(((.((((((((.(((..((((((((.......)))))))))))...)))))...((((((((....)))...))))).............)).).)))))))... ( -38.70) >sp_ag.0 22124 120 + 2160267/360-480 ACUGUGAGAUGGACCUGCGUUGUAUUAGCUAGUUGGUGAGGUAAAGGCUCACCAAGGCGACGAUACAUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGAC ..((((..............((((((.(((..((((((((.......)))))))))))...))))))..((((((((....)))...)))))))))(((((.(((.....)))))))).. ( -44.50) >sp_pn.0 341324 120 - 2038615/360-480 ACUACCAGAUGGACCUGCGUUGUAUUAGCUAGUUGGUGGGGUAACGGCUCACCAAGGCGACGAUACAUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGAC ...(((((......))).))((((((.(((..((((((((.......)))))))))))...))))))..((((((((....)))...)))))....(((((.(((.....)))))))).. ( -42.10) >sp_mu.0 152460 120 - 2030921/360-480 ACUAGUAGAUGGACCUGCGUUGUAUUAGCUAGUUGGUAAGGUAAGAGCUUACCAAGGCGACGAUACAUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGAC ....((((......))))..((((((.(((..((((((((.......)))))))))))...))))))..((((((((....)))...)))))....(((((.(((.....)))))))).. ( -42.50) >consensus ACUUAUAGAUGGACCCGCGCUGUAUUAGCUAGUUGGUAAGGUAACGGCUUACCAAGGCGACGAUACAUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGAACUGAGACACGGCCCAGAC .........(((.((.((..((((((.(((..((((((((.......)))))))))))...))))))..))..((((....))))....)))))..(((((.(((.....)))))))).. (-39.81 = -40.37 + 0.56)

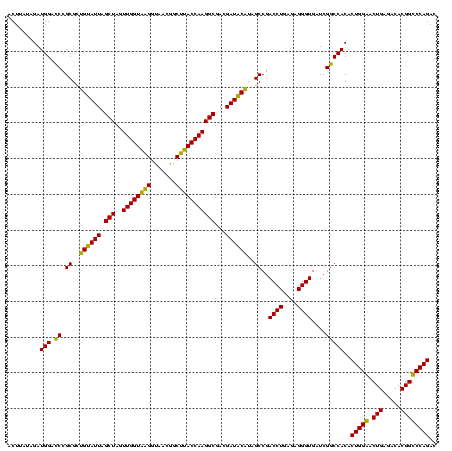
| Location | 184,579 – 184,699 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.28 |
| Mean single sequence MFE | -40.48 |
| Consensus MFE | -39.44 |
| Energy contribution | -38.33 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 184579 120 - 2685015/360-480 GUCUGGACCGUGUCUCAGUUCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGUAUCGUCGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUACGGCGCGGGUCCAUCUAUAAGU ...(((((((((((...........((((((((((.........))))))))))((((....((.((((((((.......))))))))...))..))))))))).))))))......... ( -43.90) >sy_sa.0 204973 120 - 2516575/360-480 GUCUGGACCGUGUCUCAGUUCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGUAUCGUCGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUACGGCGCGGGUCCAUCUAUAAGU ...(((((((((((...........((((((((((.........))))))))))((((....((.((((((((.......))))))))...))..))))))))).))))))......... ( -43.90) >sy_au.0 572678 120 + 2809422/360-480 GUCUGGACCGUGUCUCAGUUCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUAUGCAUCGUUGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUGCAGCGCGGAUCCAUCUAUAAGU ...((((((((((..((......((((((((((((.........))))))))))))...((((..((((((((.......)))))))).))))...))..)))))).))))......... ( -42.40) >sp_ag.0 22124 120 + 2160267/360-480 GUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUAUGUAUCGUCGCCUUGGUGAGCCUUUACCUCACCAACUAGCUAAUACAACGCAGGUCCAUCUCACAGU ...((((((((((............((((((((((.........))))))))))((((....((.((((((((.......))))))))...))..)))).)))).))))))......... ( -39.80) >sp_pn.0 341324 120 - 2038615/360-480 GUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUAUGUAUCGUCGCCUUGGUGAGCCGUUACCCCACCAACUAGCUAAUACAACGCAGGUCCAUCUGGUAGU ...((((((((((............((((((((((.........))))))))))((((....((.((((((...........))))))...))..)))).)))).))))))......... ( -35.40) >sp_mu.0 152460 120 - 2030921/360-480 GUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUAUGUAUCGUCGCCUUGGUAAGCUCUUACCUUACCAACUAGCUAAUACAACGCAGGUCCAUCUACUAGU ...((((((((((............((((((((((.........))))))))))((((....((.((((((((.......))))))))...))..)))).)))).))))))......... ( -37.50) >consensus GUCUGGACCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUAUGUAUCGUCGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUACAACGCAGGUCCAUCUAUAAGU ...((((((((((............((((((((((.........))))))))))((((....((.((((((((.......))))))))...))..)))).)))).))))))......... (-39.44 = -38.33 + -1.11) # Strand winner: forward (0.56)
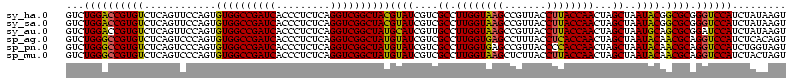
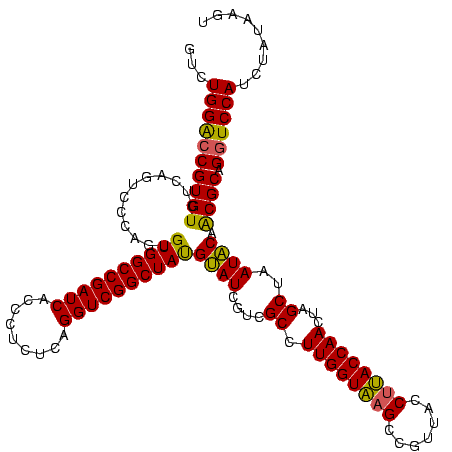
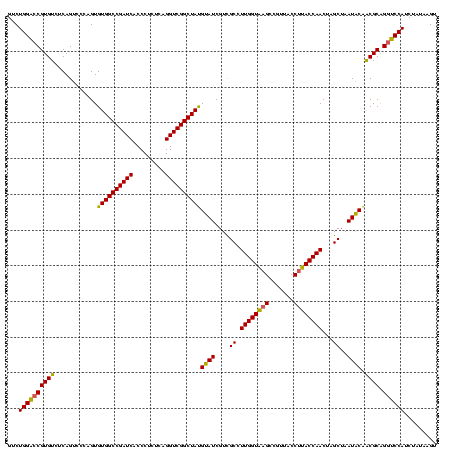
| Location | 184,579 – 184,699 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.20 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -30.73 |
| Energy contribution | -27.24 |
| Covariance contribution | -3.49 |
| Combinations/Pair | 1.54 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 184579 120 - 2685015/720-840 GCGUAGGCGGUUUUUUAAGUCUGAUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGUAAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUG ((....))(((((((............)))))))(((.(((..(((((((((..(((((.....(((((.........)))))........)))))..))))))).))))).)))..... ( -30.52) >sy_sa.0 204973 120 - 2516575/720-840 GCGUAGGCGGUUUCUUAAGUCUGAUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGGAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUG (((((..((((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))).))..)))...........((((....)))).....))......... ( -33.70) >sy_au.0 572678 120 + 2809422/720-840 GCGUAGGCGGUUUUUUAAGUCUGAUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGAAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUG (((((..((((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))).))..)))...........((((....)))).....))......... ( -31.40) >sp_ag.0 22124 119 + 2160267/720-840 GCGCAGGCGGUUCUUUAAGUCUGAAGUUAAAGGCAGUGGCUUAACCAUUGUACGCU-UUGGAAACUGGAGGACUUGAGUGCAGAAGGGGAGAGUGGAAUUCCAUGUGUAGCGGUGAAAUG (((((..((((((((((..((..((((.....(((((((.....)))))))..)))-)..))...)))))))).))..)))...........((((....)))).....))......... ( -35.60) >sp_pn.0 341324 119 - 2038615/720-840 GCGCAGGCGGUUAGAUAAGUCUGAAGUUAAAGGCUGUGGCUUAACCAUAGUAGGCU-UUGGAAACUGUUUAACUUGAGUGCAAGAGGGGAGAGUGGAAUUCCAUGUGUAGCGGUGAAAUG .(((..(((((((((((..((..(((((....(((((((.....))))))).))))-)..))...)))))))))....((((....((((........))))...)))))).)))..... ( -35.90) >sp_mu.0 152460 119 - 2030921/720-840 GCGCAGGCGGUCAGGAAAGUCUGGAGUAAAAGGCUAUGGCUCAACCAUAGUGUGCU-CUGGAAACUGUCUGACUUGAGUGCAGAAGGGGAGAGUGGAAUUCCAUGUGUAGCGGUGAAAUG (((((..((((((((....((..(((((....(((((((.....))))))).))))-)..)).....)))))).))..)))...........((((....)))).....))......... ( -40.50) >consensus GCGCAGGCGGUUUUUUAAGUCUGAAGUGAAAGCCCACGGCUCAACCAUGGAGGGCC_UUGGAAACUGGAAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUG (((((..((((((((((..((..(.........((((((.....)))))).......)..))...)))))))).))..)))...........((((....)))).....))......... (-30.73 = -27.24 + -3.49)
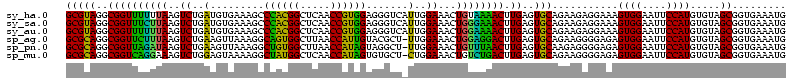
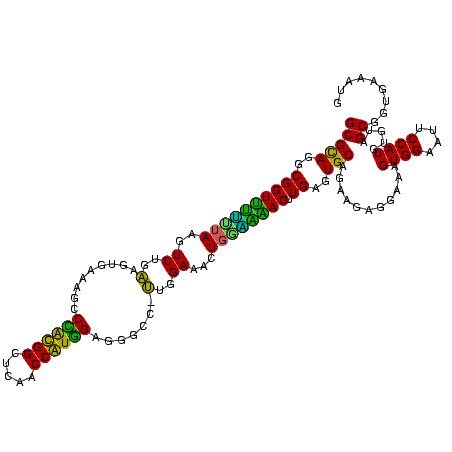
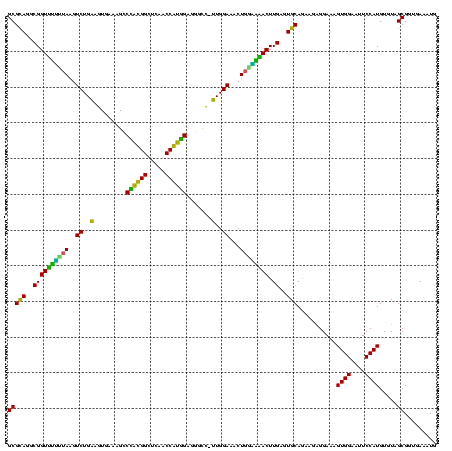
| Location | 184,579 – 184,699 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.39 |
| Mean single sequence MFE | -42.15 |
| Consensus MFE | -40.79 |
| Energy contribution | -39.23 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.10 |
| Structure conservation index | 0.97 |
| SVM decision value | 5.02 |
| SVM RNA-class probability | 0.999969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 184579 120 - 2685015/800-920 CAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACGCUGAUGUGCGAAAGCGUGGGGAU ((((..(((((.(((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).)))))))..))))...((.(((((..........))))).))... ( -40.00) >sy_sa.0 204973 120 - 2516575/800-920 CAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACGCUGAUGUGCGAAAGCGUGGGGAU ((((..(((((.(((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).)))))))..))))...((.(((((..........))))).))... ( -40.00) >sy_au.0 572678 120 + 2809422/800-920 CAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACGCUGAUGUGCGAAAGCGUGGGGAU ((((..(((((.(((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).)))))))..))))...((.(((((..........))))).))... ( -40.00) >sp_ag.0 22124 120 + 2160267/800-920 CAGAAGGGGAGAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAUAUAUGGAGGAACACCGGUGGCGAAAGCGGCUCUCUGGUCUGUAACUGACGCUGAGGCUCGAAAGCGUGGGGAG ((((.((((((.(((...((((((((((.(((......)))....))))))))))...)))).((.((....)).)))))))..))))...((.(((((..........))))).))... ( -44.20) >sp_pn.0 341324 120 - 2038615/800-920 CAAGAGGGGAGAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAUAUAUGGAGGAACACCGGUGGCGAAAGCGGCUCUCUGGCUUGUAACUGACGCUGAGGCUCGAAAGCGUGGGGAG ((((.((((((.(((...((((((((((.(((......)))....))))))))))...)))).((.((....)).)))))))..))))...((.(((((..........))))).))... ( -43.40) >sp_mu.0 152460 120 - 2030921/800-920 CAGAAGGGGAGAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAUAUAUGGAGGAACACCAGUGGCGAAAGCGGCUCUCUGGUCUGUCACUGACGCUGAGGCUCGAAAGCGUGGGUAG ((((.((((((.(((...((((((((((.(((......)))....))))))))))...)))).((.((....)).)))))))..))))..(((.(((((..........))))).))).. ( -45.30) >consensus CAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAAGCGACUCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGGGAG ((((...((((((((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).)))))))..))))...((.(((((..........))))).))... (-40.79 = -39.23 + -1.55)

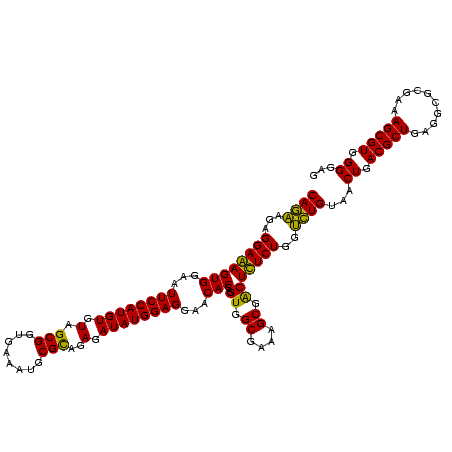
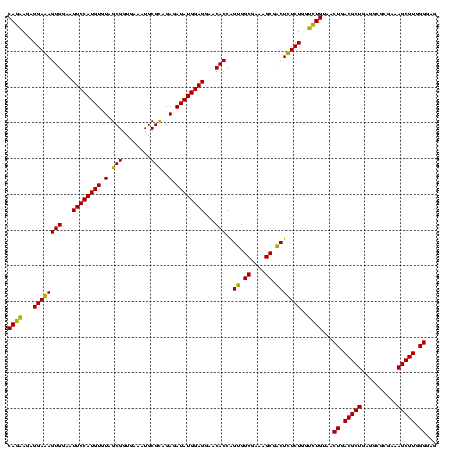
| Location | 184,579 – 184,699 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.11 |
| Mean single sequence MFE | -46.05 |
| Consensus MFE | -46.47 |
| Energy contribution | -46.05 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.32 |
| Structure conservation index | 1.01 |
| SVM decision value | 5.04 |
| SVM RNA-class probability | 0.999970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 184579 120 - 2685015/920-1040 CAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACC ....((((.........))))(((.(((..((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))....))).))).... ( -45.50) >sy_sa.0 204973 120 - 2516575/920-1040 CAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACC ....((((.........))))(((.(((..((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))....))).))).... ( -45.50) >sy_au.0 572678 120 + 2809422/920-1040 CAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACC ....((((.........))))(((.(((..((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))....))).))).... ( -45.50) >sp_ag.0 22124 120 + 2160267/920-1040 CAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGCCCUUUCCGGGGCUUAGUGCCGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACC ....((((.........))))(((.(((..((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))....))).))).... ( -47.80) >sp_pn.0 341324 120 - 2038615/920-1040 CAAACAGGAUUAGAUACCCUGGUAGUCCACGCUGUAAACGAUGAGUGCUAGGUGUUAGACCCUUUCCGGGGUUUAGUGCCGUAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACC ....((((.........))))(((.(((..((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))....))).))).... ( -44.20) >sp_mu.0 152460 120 - 2030921/920-1040 CGAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGCCCUUUCCGGGGCUUAGUGCCGGAGCUAACGCAAUAAGCACUCCGCCUGGGGAGUACGACC ....((((.........))))(((.(((..((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))....))).))).... ( -47.80) >consensus CAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGCCCUUUCCGCCCCUUAGUGCCGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACC ....((((.........))))(((.(((..((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))....))).))).... (-46.47 = -46.05 + -0.42)

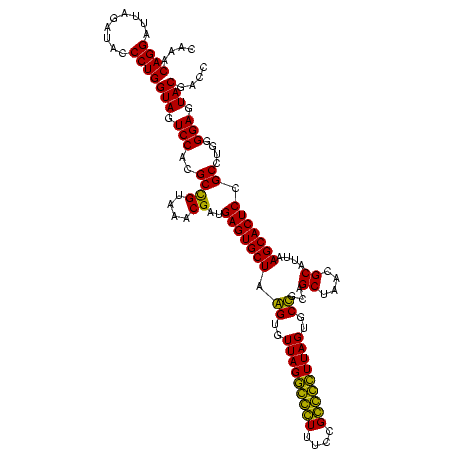

| Location | 184,579 – 184,699 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.11 |
| Mean single sequence MFE | -40.79 |
| Consensus MFE | -40.09 |
| Energy contribution | -38.48 |
| Covariance contribution | -1.61 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 184579 120 - 2685015/920-1040 GGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUG ((((.(((.((..((((((((((((....(.((((((....)).....((((.(....))))))))).)..)))))))..)))))..)).)))))))..((((((.....)))))).... ( -43.50) >sy_sa.0 204973 120 - 2516575/920-1040 GGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUG ((((.(((.((..((((((((((((....(.((((((....)).....((((.(....))))))))).)..)))))))..)))))..)).)))))))..((((((.....)))))).... ( -43.50) >sy_au.0 572678 120 + 2809422/920-1040 GGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUG ((((.(((.((..((((((((((((....(.((((((....)).....((((.(....))))))))).)..)))))))..)))))..)).)))))))..((((((.....)))))).... ( -43.50) >sp_ag.0 22124 120 + 2160267/920-1040 GGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCGGCACUAAGCCCCGGAAAGGGCCUAACACCUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUG ((((.(((.((..((((((((((((....(.(((((..(((((.....)))((.....))))))))).)..)))))))..)))))..)).)))))))..((((((.....)))))).... ( -40.00) >sp_pn.0 341324 120 - 2038615/920-1040 GGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUACGGCACUAAACCCCGGAAAGGGUCUAACACCUAGCACUCAUCGUUUACAGCGUGGACUACCAGGGUAUCUAAUCCUGUUUG ((((.(((.(...((((((((((((....(.((((((....)).....((((......)))).)))).)..)))))))..)))))...).)))))))..((((((.....)))))).... ( -35.30) >sp_mu.0 152460 120 - 2030921/920-1040 GGUCGUACUCCCCAGGCGGAGUGCUUAUUGCGUUAGCUCCGGCACUAAGCCCCGGAAAGGGCCUAACACCUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUCG ((((.(((.((..((((((((((((....(.(((((..(((((.....))).......))..))))).)..)))))))..)))))..)).)))))))..((((((.....)))))).... ( -38.91) >consensus GGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGCCCCGGAAACCCCCUAACACCUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUG ((((.(((.((..((((((((((((....(.((((((....)).....((((......)))).)))).)..)))))))..)))))..)).)))))))..((((((.....)))))).... (-40.09 = -38.48 + -1.61) # Strand winner: forward (0.91)

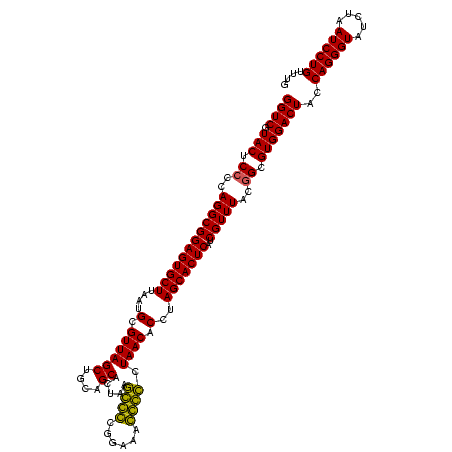
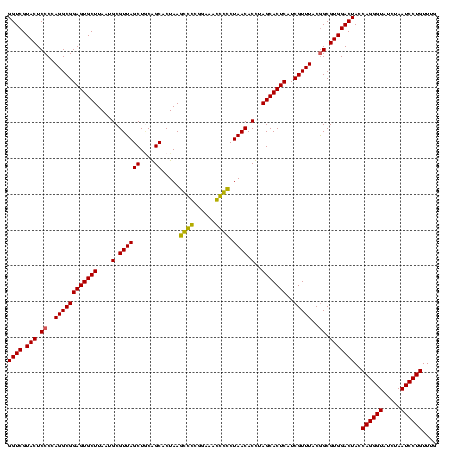
| Location | 184,579 – 184,699 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -54.65 |
| Consensus MFE | -54.89 |
| Energy contribution | -54.62 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -4.22 |
| Structure conservation index | 1.00 |
| SVM decision value | 5.92 |
| SVM RNA-class probability | 0.999995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 184579 120 - 2685015/960-1080 AUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACA ..(((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..)))...((.(((....))))).. ( -53.80) >sy_sa.0 204973 120 - 2516575/960-1080 AUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACA ..(((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..)))...((.(((....))))).. ( -53.80) >sy_au.0 572678 120 + 2809422/960-1080 AUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACA ..(((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..)))...((.(((....))))).. ( -53.80) >sp_ag.0 22124 120 + 2160267/960-1080 AUGAGUGCUAGGUGUUAGGCCCUUUCCGGGGCUUAGUGCCGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCACA ..(((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..)))...((.(((....))))).. ( -56.10) >sp_pn.0 341324 120 - 2038615/960-1080 AUGAGUGCUAGGUGUUAGACCCUUUCCGGGGUUUAGUGCCGUAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCACA ..(((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..)))...((.(((....))))).. ( -54.10) >sp_mu.0 152460 120 - 2030921/960-1080 AUGAGUGCUAGGUGUUAGGCCCUUUCCGGGGCUUAGUGCCGGAGCUAACGCAAUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCACA ....((((..((..(((((((((....)))))))))..))((.((....)).....((.((((((((...((((.(((((....)))))..))))..)))......))))))))))))). ( -56.30) >consensus AUGAGUGCUAAGUGUUAGGCCCUUUCCGCCCCUUAGUGCCGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACA ..(((((((.((..(((((((((....)))))))))..))...((....))....)))))))..(((...((((.(((((....)))))..))))..)))...((.(((....))))).. (-54.89 = -54.62 + -0.28)

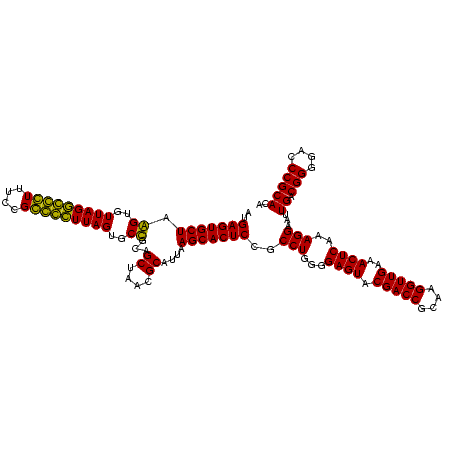
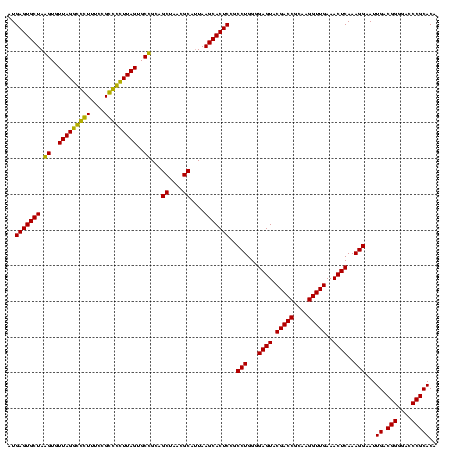
| Location | 184,579 – 184,699 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -39.27 |
| Consensus MFE | -37.21 |
| Energy contribution | -35.43 |
| Covariance contribution | -1.78 |
| Combinations/Pair | 1.11 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 184579 120 - 2685015/960-1080 UGUGCGGGUCCCCGUCAAUUCCUUUGAGUUUCAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAU ((.((((....))))))...(((..((((..(.(((....))).).))))...)))..(((((((....(.((((((....)).....((((.(....))))))))).)..))))))).. ( -39.70) >sy_sa.0 204973 120 - 2516575/960-1080 UGUGCGGGUCCCCGUCAAUUCCUUUGAGUUUCAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAU ((.((((....))))))...(((..((((..(.(((....))).).))))...)))..(((((((....(.((((((....)).....((((.(....))))))))).)..))))))).. ( -39.70) >sy_au.0 572678 120 + 2809422/960-1080 UGUGCGGGUCCCCGUCAAUUCCUUUGAGUUUCAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAU ((.((((....))))))...(((..((((..(.(((....))).).))))...)))..(((((((....(.((((((....)).....((((.(....))))))))).)..))))))).. ( -39.70) >sp_ag.0 22124 120 + 2160267/960-1080 UGUGCGGGCCCCCGUCAAUUCCUUUGAGUUUCAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCGGCACUAAGCCCCGGAAAGGGCCUAACACCUAGCACUCAU .(((((((((((((...........((((..(.(((....))).).))))....(((..((((((....((....))...))))))..))).)))...))))))........)))).... ( -40.50) >sp_pn.0 341324 120 - 2038615/960-1080 UGUGCGGGCCCCCGUCAAUUCCUUUGAGUUUCAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUACGGCACUAAACCCCGGAAAGGGUCUAACACCUAGCACUCAU .((((((....))((....((((((((((..(.(((....))).).)))).((.((.(.((((((....((....))...))))))...))).))))))))....)).....)))).... ( -35.50) >sp_mu.0 152460 120 - 2030921/960-1080 UGUGCGGGCCCCCGUCAAUUCCUUUGAGUUUCAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAUUGCGUUAGCUCCGGCACUAAGCCCCGGAAAGGGCCUAACACCUAGCACUCAU .(((((((((((((...........((((..(.(((....))).).))))....(((..((((((....((....))...))))))..))).)))...))))))........)))).... ( -40.50) >consensus UGUGCGGGCCCCCGUCAAUUCCUUUGAGUUUCAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGCCCCGGAAACCCCCUAACACCUAGCACUCAU ((.((((....))))))...(((..((((..(.(((....))).).))))...)))..(((((((....(.((((((....)).....((((......)))).)))).)..))))))).. (-37.21 = -35.43 + -1.78) # Strand winner: forward (0.97)

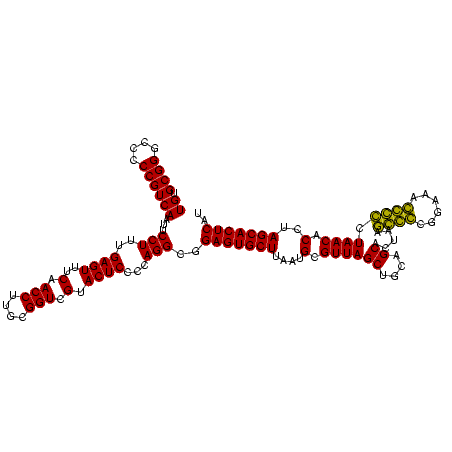

| Location | 184,579 – 184,699 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.72 |
| Mean single sequence MFE | -39.98 |
| Consensus MFE | -38.90 |
| Energy contribution | -39.07 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 184579 120 - 2685015/1000-1120 GCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGA ((.(((..((.(((((((.(((((((.(((((((.(((((....)))))..))))...(.......).....)))((....)))))))))......))))))).)).)))...))..... ( -39.80) >sy_sa.0 204973 120 - 2516575/1000-1120 GCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGA ((.(((..((.(((((((.(((((((.(((((((.(((((....)))))..))))...(.......).....)))((....)))))))))......))))))).)).)))...))..... ( -39.80) >sy_au.0 572678 120 + 2809422/1000-1120 GCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGA ((.(((..((.(((((((.(((((((.(((((((.(((((....)))))..))))...(.......).....)))((....)))))))))......))))))).)).)))...))..... ( -39.80) >sp_ag.0 22124 120 + 2160267/1000-1120 GCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGA ((.(((..((.(((((((.((((((((...((((.(((((....)))))..))))..)))......)))))((((((.......)))).)).....))))))).)).)))...))..... ( -40.30) >sp_pn.0 341324 120 - 2038615/1000-1120 GUAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGA ((.(((..((.(((((((.((((((((...((((.(((((....)))))..))))..)))......)))))((((((.......)))).)).....))))))).)).))).))....... ( -40.70) >sp_mu.0 152460 120 - 2030921/1000-1120 GGAGCUAACGCAAUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGA .((((((..((.....((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....))))....))...))))))..((((........)))).. ( -39.50) >consensus GCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGA ...(((..((.(((((((.((((((((...((((.(((((....)))))..))))..)))......))))).....((((.((......)).))))))))))).)).))).......... (-38.90 = -39.07 + 0.17)

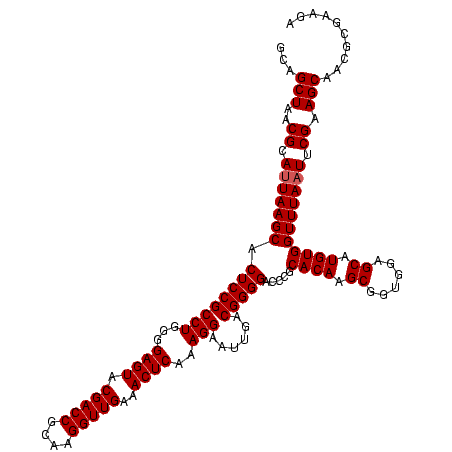
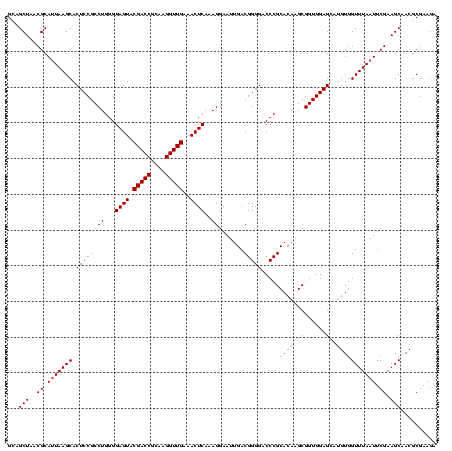
| Location | 184,579 – 184,699 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -42.70 |
| Consensus MFE | -41.42 |
| Energy contribution | -42.70 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 184579 120 - 2685015/1200-1320 AUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAAGUUGACUGCCGGUGACAAACCGG ..((((((.....(((((((((....)))((.(((......))))).))))))))))))....(((((((.((((..(.....)...))))..))))))).....(((((.....))))) ( -43.30) >sy_sa.0 204973 120 - 2516575/1200-1320 AUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAGGUUGACUGCCGGUGACAAACCGG ..((((((.....(((((((((....)))((.(((......))))).))))))))))))....(((((((.((((..(.....)...))))..))))))).....(((((.....))))) ( -43.00) >sy_au.0 572678 120 + 2809422/1200-1320 AUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAAGUUGACUGCCGGUGACAAACCGG ..((((((.....(((((((((....)))((.(((......))))).))))))))))))....(((((((.((((..(.....)...))))..))))))).....(((((.....))))) ( -43.30) >sp_ag.0 22124 120 + 2160267/1200-1320 AUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCCUAUUGUUAGUUGCCAUCAUUAAGUUGGGCACUCUAGCGAGACUGCCGGUAAUAAACCGG ..((((((.....(((((((((....)))((.(((......))))).))))))))))))....(((((((.((((..(.....)...))))..))))))).....(((((.....))))) ( -42.20) >sp_pn.0 341324 120 - 2038615/1200-1320 AUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCCUAUUGUUAGUUGCCAUCAUUCAGUUGGGCACUCUAGCGAGACUGCCGGUAAUAAACCGG ..((((((.....(((((((((....)))((.(((......))))).))))))))))))....(((((((.((((..(.....)...))))..))))))).....(((((.....))))) ( -42.20) >sp_mu.0 152460 120 - 2030921/1200-1320 AUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUAUUGUUAGUUGCCAUCAUUAAGUUGGGCACUCUAGCGAGACUGCCGGUAAUAAACCGG ..((((((.....(((((((((....)))((.(((......))))).))))))))))))....(((((((.((((..(.....)...))))..))))))).....(((((.....))))) ( -42.20) >consensus AUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAGCGAGACUGCCGGUAACAAACCGG ..((((((.....(((((((((....)))((.(((......))))).))))))))))))....(((((((.((((..(.....)...))))..))))))).....(((((.....))))) (-41.42 = -42.70 + 1.28)

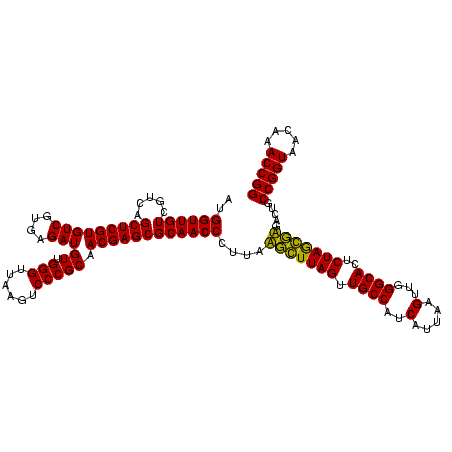
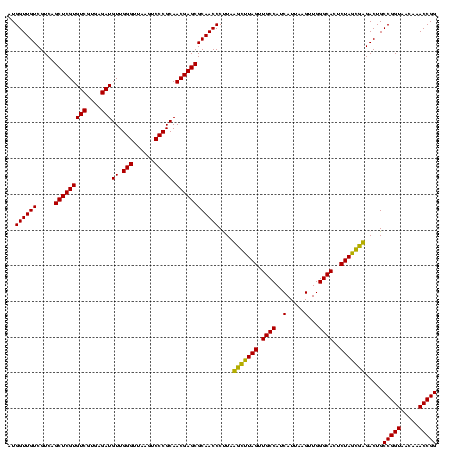
| Location | 184,579 – 184,699 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -37.03 |
| Consensus MFE | -37.92 |
| Energy contribution | -36.20 |
| Covariance contribution | -1.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.76 |
| Structure conservation index | 1.02 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870183 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 184579 120 - 2685015/1240-1360 UCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAAGUUGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAUG ...(((.....)))....((((.(((((((.((((..(.....)...))))..)))))))..((.(((((.....))))).)))))).((((((((.(......).)))).))))..... ( -37.60) >sy_sa.0 204973 120 - 2516575/1240-1360 UCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAGGUUGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAUG ...(((.....)))....((((.(((((((.((((..(.....)...))))..)))))))..((.(((((.....))))).)))))).((((((((.(......).)))).))))..... ( -37.30) >sy_au.0 572678 120 + 2809422/1240-1360 UCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAAGUUGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAUG ...(((.....)))....((((.(((((((.((((..(.....)...))))..)))))))..((.(((((.....))))).)))))).((((((((.(......).)))).))))..... ( -37.60) >sp_ag.0 22124 120 + 2160267/1240-1360 UCCCGCAACGAGCGCAACCCCUAUUGUUAGUUGCCAUCAUUAAGUUGGGCACUCUAGCGAGACUGCCGGUAAUAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAUG ...(((.....)))..((((((.(((((((.((((..(.....)...))))..))))))).....(((((.....))))))))..)))((((((((.(......).)))).))))..... ( -36.60) >sp_pn.0 341324 120 - 2038615/1240-1360 UCCCGCAACGAGCGCAACCCCUAUUGUUAGUUGCCAUCAUUCAGUUGGGCACUCUAGCGAGACUGCCGGUAAUAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAUG ...(((.....)))..((((((.(((((((.((((..(.....)...))))..))))))).....(((((.....))))))))..)))((((((((.(......).)))).))))..... ( -36.60) >sp_mu.0 152460 120 - 2030921/1240-1360 UCCCGCAACGAGCGCAACCCUUAUUGUUAGUUGCCAUCAUUAAGUUGGGCACUCUAGCGAGACUGCCGGUAAUAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAUG ...(((.....)))....((((.(((((((.((((..(.....)...))))..)))))))..((.(((((.....))))).)))))).((((((((.(......).)))).))))..... ( -36.50) >consensus UCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAGCGAGACUGCCGGUAACAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAUG ...(((.....)))..(((....(((((((.((((..(.....)...))))..)))))))..((.(((((.....))))).))..)))((((((((.(......).)))).))))..... (-37.92 = -36.20 + -1.72)


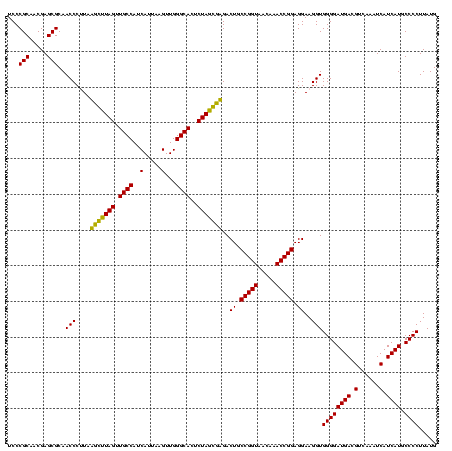
| Location | 184,579 – 184,699 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -42.45 |
| Consensus MFE | -38.18 |
| Energy contribution | -38.18 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988481 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 184579 120 - 2685015/1240-1360 CAUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCAACUUAGAGUGCCCAACUUAAUGAUGGCAACUAAGCUUAAGGGUUGCGCUCGUUGCGGGA .....((((.((((((......))))))))))....(((((((((.....)))))..(.((((...((((((....(((((.(..(....)....))))))....))))))))))))))) ( -39.20) >sy_sa.0 204973 120 - 2516575/1240-1360 CAUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCAACCUAGAGUGCCCAACUUAAUGAUGGCAACUAAGCUUAAGGGUUGCGCUCGUUGCGGGA .....((((.((((((......))))))))))....(((((((((.....)))))..(.((((...((((((....(((((.(..(....)....))))))....))))))))))))))) ( -39.20) >sy_au.0 572678 120 + 2809422/1240-1360 CAUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCAACUUAGAGUGCCCAACUUAAUGAUGGCAACUAAGCUUAAGGGUUGCGCUCGUUGCGGGA .....((((.((((((......))))))))))....(((((((((.....)))))..(.((((...((((((....(((((.(..(....)....))))))....))))))))))))))) ( -39.20) >sp_ag.0 22124 120 + 2160267/1240-1360 CAUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUAUUACCGGCAGUCUCGCUAGAGUGCCCAACUUAAUGAUGGCAACUAACAAUAGGGGUUGCGCUCGUUGCGGGA .....((((.((((((......))))))))))......(((((((.....))))).))((((((..((((((....((((.((..(....)...)).))))....))))))...)))))) ( -46.20) >sp_pn.0 341324 120 - 2038615/1240-1360 CAUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUAUUACCGGCAGUCUCGCUAGAGUGCCCAACUGAAUGAUGGCAACUAACAAUAGGGGUUGCGCUCGUUGCGGGA .....((((.((((((......))))))))))......(((((((.....))))).))((((((..((((((.(..(((......(....)......)))..)..))))))...)))))) ( -44.40) >sp_mu.0 152460 120 - 2030921/1240-1360 CAUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUAUUACCGGCAGUCUCGCUAGAGUGCCCAACUUAAUGAUGGCAACUAACAAUAAGGGUUGCGCUCGUUGCGGGA .....((((.((((((......))))))))))......(((((((.....))))).))((((((..((((((....((((.((..(....)...)).))))....))))))...)))))) ( -46.50) >consensus CAUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUAUCACCGGCAGUCAACCUAGAGUGCCCAACUUAAUGAUGGCAACUAACAAUAAGGGUUGCGCUCGUUGCGGGA .....((((.((((((......))))))))))....((((.((((.....)))((((.((......)).))))......(((((..((((((.........))))))..)))))).)))) (-38.18 = -38.18 + -0.00) # Strand winner: reverse (0.71)


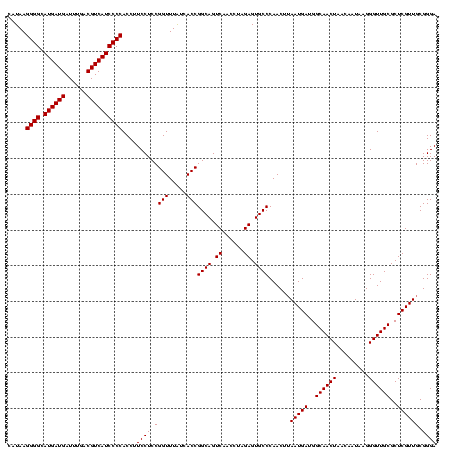
| Location | 184,579 – 184,699 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.06 |
| Mean single sequence MFE | -36.67 |
| Consensus MFE | -36.96 |
| Energy contribution | -33.88 |
| Covariance contribution | -3.08 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.40 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.902277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 184579 120 - 2685015/1360-1480 AUUUGGGCUACACACGUGCUACAAUGGACAAUACAAAGGGCAGCGAAACCGCGAGGUCAAGCAAAUCCCAUAAAGUUGUUCUCAGUUCGGAUUGUAGUCUGCAACUCGACUACAUGAAGC (((((((((.((....)).......(((((((.....(((..((...(((....)))...))....))).....)))))))..)))))))))(((((((........)))))))...... ( -31.10) >sy_sa.0 204973 120 - 2516575/1360-1480 AUUUGGGCUACACACGUGCUACAAUGGACAAUACAAAGGGCAGCUAAACCGCGAGGUCAUGCAAAUCCCAUAAAGUUGUUCUCAGUUCGGAUUGUAGUCUGCAACUCGACUACAUGAAGC (((((((((.((....)).......(((((((.....(((..((...(((....)))...))....))).....)))))))..)))))))))(((((((........)))))))...... ( -30.70) >sy_au.0 572678 120 + 2809422/1360-1480 AUUUGGGCUACACACGUGCUACAAUGGACAAUACAAAGGGCAGCGAAACCGCGAGGUCAAGCAAAUCCCAUAAAGUUGUUCUCAGUUCGGAUUGUAGUCUGCAACUCGACUACAUGAAGC (((((((((.((....)).......(((((((.....(((..((...(((....)))...))....))).....)))))))..)))))))))(((((((........)))))))...... ( -31.10) >sp_ag.0 22124 120 + 2160267/1360-1480 ACCUGGGCUACACACGUGCUACAAUGGUUGGUACAACGAGUCGCAAGCCGGUGACGGCAAGCUAAUCUCUUAAAGCCAAUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAGU ..(((((((.((....)).......(((((((.....(((..((..((((....))))..))....))).....)))))))..)))))))..(((((((........)))))))...... ( -39.70) >sp_pn.0 341324 120 - 2038615/1360-1480 ACCUGGGCUACACACGUGCUACAAUGGCUGGUACAACGAGUCGCAAGCCGGUGACGGCAAGCUAAUCUCUUAAAGCCAGUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAGU ..(((((((.((....)).......(((((((.....(((..((..((((....))))..))....))).....)))))))..)))))))..(((((((........)))))))...... ( -42.00) >sp_mu.0 152460 120 - 2030921/1360-1480 ACCUGGGCUACACACGUGCUACAAUGGUCGGUACAACGAGUUGCGAGCCGGUGACGGCAAGCUAAUCUCUGAAAGCCGAUCUCAGUUCGGAUUGGAGGCUGCAACUCGCCUCCAUGAAGU ..(((((((.((....)).......(((((((.....(((..((..((((....))))..))....))).....)))))))..)))))))..(((((((........)))))))...... ( -45.40) >consensus ACCUGGGCUACACACGUGCUACAAUGGACAAUACAAAGAGCAGCGAAACCGCGACGGCAAGCAAAUCCCAUAAAGCCGUUCUCAGUUCGGAUUGUAGGCUGCAACUCGACUACAUGAAGC ..(((((((.((....)).......(((((((.....(((..((...(((....)))...))....))).....)))))))..)))))))..(((((((........)))))))...... (-36.96 = -33.88 + -3.08)
| Location | 184,579 – 184,697 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.18 |
| Mean single sequence MFE | -42.72 |
| Consensus MFE | -41.87 |
| Energy contribution | -40.48 |
| Covariance contribution | -1.39 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 184579 118 - 2685015/1520-1640 UACGUUCCCGGGUCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGCCGGUGGAGUAACCAUUU--GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAA ........(((((..((....))..)))))((((.((.(((..((((((..(((((((...((((..(((..((....--)).))).)))))))..))))..))))))..))))))))). ( -41.50) >sy_sa.0 204973 120 - 2516575/1520-1640 UACGUUCCCGGGUCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGCCGGUGGAGUAACCAUUUAUGGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAA ........(((((..((....))..)))))((((.((.(((..((((((..(((((((...((((..(((..(((....))).))).)))))))..))))..))))))..))))))))). ( -42.10) >sy_au.0 572678 119 + 2809422/1520-1640 UACGUUCCCGGGUCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGCCGGUGGAGUAACCUUUUA-GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAA ........(((((..((....))..)))))((((.((.(((..((((((..(((((((...((((..(((..((.....-)).))).)))))))..))))..))))))..))))))))). ( -41.40) >sp_ag.0 22124 119 + 2160267/1520-1640 UACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGUCGGUGAGGUAACCUUUUA-GGAGCCAGCCGCCUAAGGUGGGAUAGAUGAUUGGGGUGAA ........(((((..((....))..)))))((((.((.(((..((((((..((((...((.((((..(((..((.....-)).))).)))).))..))))..))))))..))))))))). ( -42.80) >sp_pn.0 341324 118 - 2038615/1520-1640 UACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGUCGGUGAGGUAACCGUA-A-GGAGCCAGCCGCCUAAGGUGGGAUAGAUGAUUGGGGUGAA ........(((((..((....))..)))))((((.((.(((..((((((..((((...((.((((..(((..((...-.-)).))).)))).))..))))..))))))..))))))))). ( -45.30) >sp_mu.0 152460 119 - 2030921/1520-1640 UACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGUCGGUGAGGUAACCUUUUA-GGGGCCAGCCGCCUAAGGUGGGAUGGAUGAUUGGGGUGAA ........(((((..((....))..)))))((((.((.(((..(((..(..((((...((.((((..(((..(((....-)))))).)))).))..))))..)..)))..))))))))). ( -43.20) >consensus UACGUUCCCGGGCCUUGUACACACCGCCCGUCACACCACGAGAGUUUGUAACACCCGAAGCCGGUGAAGUAACCUUUUA_GGAGCCAGCCGCCGAAGGUGGGACAAAUGAUUGGGGUGAA ........(((((..((....))..)))))((((.((.(((..((((((..((((....(.((((..(((..((......)).))).)))).)...))))..))))))..))))))))). (-41.87 = -40.48 + -1.39)

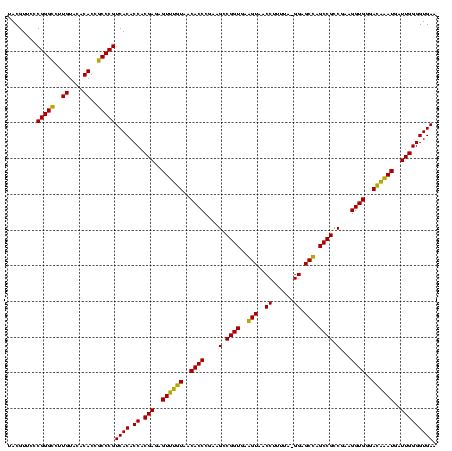
| Location | 184,579 – 184,697 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.69 |
| Mean single sequence MFE | -43.07 |
| Consensus MFE | -41.67 |
| Energy contribution | -40.53 |
| Covariance contribution | -1.14 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.79 |
| SVM RNA-class probability | 0.999950 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 184579 118 - 2685015/1560-1680 AGAGUUUGUAACACCCGAAGCCGGUGGAGUAACCAUUU--GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUC ...((((((..(((((((...((((..(((..((....--)).))).)))))))..))))..))))))((((.......)))).........((((((((((....)))))))))).... ( -42.30) >sy_sa.0 204973 120 - 2516575/1560-1680 AGAGUUUGUAACACCCGAAGCCGGUGGAGUAACCAUUUAUGGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUC ...((((((..(((((((...((((..(((..(((....))).))).)))))))..))))..))))))((((.......)))).........((((((((((....)))))))))).... ( -42.90) >sy_au.0 572678 119 + 2809422/1560-1680 AGAGUUUGUAACACCCGAAGCCGGUGGAGUAACCUUUUA-GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUC ...((((((..(((((((...((((..(((..((.....-)).))).)))))))..))))..))))))((((.......)))).........((((((((((....)))))))))).... ( -42.20) >sp_ag.0 22124 119 + 2160267/1560-1680 AGAGUUUGUAACACCCGAAGUCGGUGAGGUAACCUUUUA-GGAGCCAGCCGCCUAAGGUGGGAUAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUC ...((((((..((((...((.((((..(((..((.....-)).))).)))).))..))))..))))))((((.......)))).........((((((((((....)))))))))).... ( -42.10) >sp_pn.0 341324 118 - 2038615/1560-1680 AGAGUUUGUAACACCCGAAGUCGGUGAGGUAACCGUA-A-GGAGCCAGCCGCCUAAGGUGGGAUAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUC ...((((((..((((...((.((((..(((..((...-.-)).))).)))).))..))))..))))))((((.......)))).........((((((((((....)))))))))).... ( -44.60) >sp_mu.0 152460 119 - 2030921/1560-1680 AGAGUUUGUAACACCCGAAGUCGGUGAGGUAACCUUUUA-GGGGCCAGCCGCCUAAGGUGGGAUGGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUC ...(((((...(((((..(((((((......))).....-....(((.(((((...)))))..)))..)))).)))))....)).)))....((((((((((....)))))))))).... ( -44.30) >consensus AGAGUUUGUAACACCCGAAGCCGGUGAAGUAACCUUUUA_GGAGCCAGCCGCCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUC ...((((((..((((....(.((((..(((..((......)).))).)))).)...))))..))))))((((.......)))).........((((((((((....)))))))))).... (-41.67 = -40.53 + -1.14)

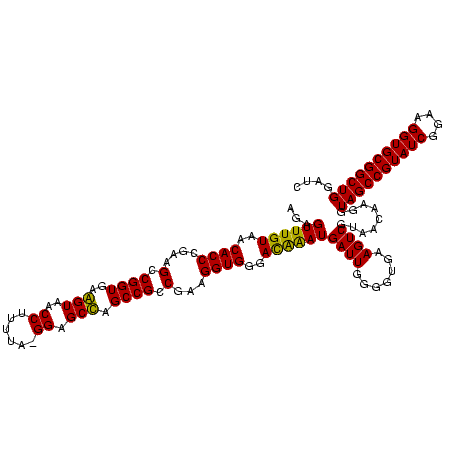
| Location | 184,579 – 184,696 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.01 |
| Mean single sequence MFE | -39.12 |
| Consensus MFE | -35.15 |
| Energy contribution | -34.90 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 184579 117 - 2685015/1600-1720 GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUCGU---AG .(((....(((.....(((((..(..((((((.......))))))........(((((((((....))))))))))..)))))((((.....))))......))).....)))..---.. ( -38.40) >sy_sa.0 204973 120 - 2516575/1600-1720 GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUUCUUACGA .........(((.(((((((...(....((..(((((((.((....))....((((((((((....))))))))))..))))).))..))....((.....)))...)))))))..))). ( -40.30) >sy_au.0 572678 120 + 2809422/1600-1720 GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUUCUUCAGA ((......))(..(((((((...(....((..(((((((.((....))....((((((((((....))))))))))..))))).))..))....((.....)))...)))))))..)... ( -39.80) >sp_ag.0 22124 99 + 2160267/1600-1720 GGAGCCAGCCGCCUAAGGUGGGAUAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUA---------------------A ....((..(((((...)))))..((((.....(((((((.((....))....((((((((((....))))))))))..)))))))...)))).))...---------------------. ( -38.30) >sp_pn.0 341324 99 - 2038615/1600-1720 GGAGCCAGCCGCCUAAGGUGGGAUAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUA---------------------A ....((..(((((...)))))..((((.....(((((((.((....))....((((((((((....))))))))))..)))))))...)))).))...---------------------. ( -38.30) >sp_mu.0 152460 99 - 2030921/1600-1720 GGGGCCAGCCGCCUAAGGUGGGAUGGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAAA---------------------A ....(((.(((((...)))))..)))..((..(((((((.((....))....((((((((((....))))))))))..))))).))..))........---------------------. ( -39.60) >consensus GGAGCCAGCCGCCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUA_____________________A ....((.(((......))).))......((..(((((((.((....))....((((((((((....))))))))))..))))).))..)).............................. (-35.15 = -34.90 + -0.25)

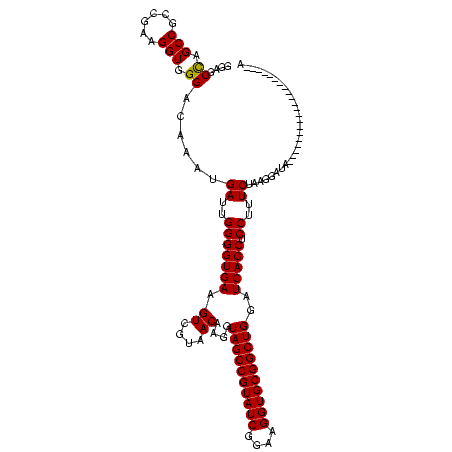
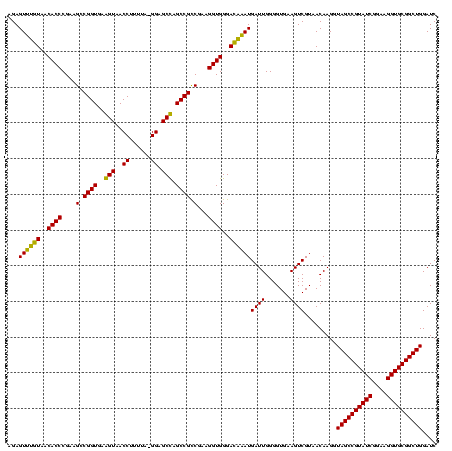
| Location | 184,579 – 184,694 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.47 |
| Mean single sequence MFE | -33.48 |
| Consensus MFE | -23.72 |
| Energy contribution | -23.88 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.71 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
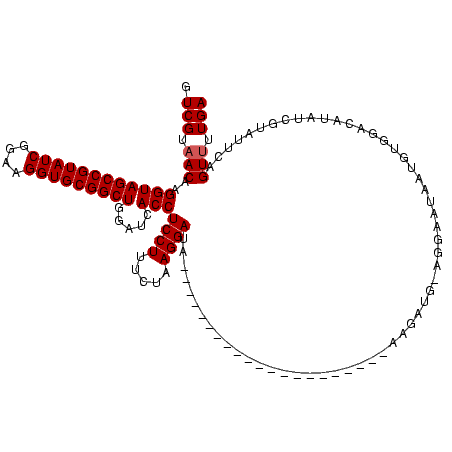
>sy_ha.0 184579 115 - 2685015/1640-1760 GUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUCGU---AGAGAUG-AGGAAUAACGU-GACAUAUUGUAUUCAGUUUUGA .(((.(((....((((((((((....))))))))))..((((.((((.....))))..(((((....((((((..---.))))))-..)))))..))-)).............))).))) ( -34.80) >sy_sa.0 204973 120 - 2516575/1640-1760 GUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUUCUUACGAAGAUGCAGGAAUAACAUUGACAUAUUGUAUUCAGUUUUGA ..........((((((((((((....))))))))).....)))((((.....)))).....(((((((((((((....))))))....(((((.((.........))))))).))))))) ( -35.30) >sy_au.0 572678 118 + 2809422/1640-1760 GUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUUCUUCAGAAGAUG-CGGAAUAAUGU-GACAUAUUGUAUUCAGUUUUGA .(((.(((....((((((((((....))))))))))..((((.((((.....))))..(((((.(..(((((((....)))))))-).)))))..))-)).............))).))) ( -38.80) >sp_ag.0 22124 93 + 2160267/1640-1760 GUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUA---------------------AGGAAACCUGCCAUUUGCG------UCUUGUUUAGUUUUGA (.(((((..(((((((((((((....)))))))))........(((((((....)).)---------------------)))).)))).....)))))------.).............. ( -30.60) >sp_pn.0 341324 90 - 2038615/1640-1760 GUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUA---------------------AGGAA--CUGCGCAUUG-G------UCUUGUUUAGUCUUGA .......((((.((((((((((....)))))))))).......(((((((....)).)---------------------))))(--(((.(((...-.------...))).)))))))). ( -29.90) >sp_mu.0 152460 98 - 2030921/1640-1760 GUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAAA---------------------AACGCA-AAGCACAUUAGGGGAACUUCUUGUUUAGUUUUGA ..(((.....((((((((((((....))))))))).....)))((((.....))))..---------------------.)))((-(((((((..(((....)))..)))...)))))). ( -31.50) >consensus GUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUA_____________________AAGAUG_AGGAAUAAUGUGGACAUAUCGUAUUCAGUUUUGA .(((.(((..((((((((((((....))))))))).....)))((((.....)))).........................................................))).))) (-23.72 = -23.88 + 0.17)

| Location | 184,579 – 184,694 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.20 |
| Mean single sequence MFE | -35.50 |
| Consensus MFE | -21.83 |
| Energy contribution | -21.83 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.88 |
| SVM RNA-class probability | 0.997525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
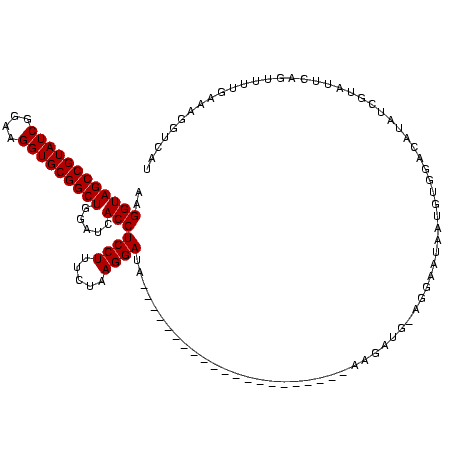
>sy_ha.0 184579 115 - 2685015/1648-1768 AAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUCGU---AGAGAUG-AGGAAUAACGU-GACAUAUUGUAUUCAGUUUUGAAUGUUUAU ..((((((((((((....))))))))).....)))((((.....)))).(((((((((((..(((((---....)))-))(((((.((.-.......))))))).))))))))))).... ( -38.40) >sy_sa.0 204973 120 - 2516575/1648-1768 AAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUUCUUACGAAGAUGCAGGAAUAACAUUGACAUAUUGUAUUCAGUUUUGAAUGCUCAU ..((((((((((((....))))))))).....)))((((.....))))..((((((((((((((((....))))))....(((((.((.........))))))).))))))))))..... ( -38.90) >sy_au.0 572678 118 + 2809422/1648-1768 AAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUUCUUCAGAAGAUG-CGGAAUAAUGU-GACAUAUUGUAUUCAGUUUUGAAUGUUUAU ..((((((((((((....))))))))).....)))((((.....)))).(((((((((((((((((....)))))).-.((((((((((-...))))))).))).))))))))))).... ( -40.00) >sp_ag.0 22124 93 + 2160267/1648-1768 AAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUA---------------------AGGAAACCUGCCAUUUGCG------UCUUGUUUAGUUUUGAGAGGUCUU .(((((((((((((....)))))))))........(((((((....)).)---------------------)))).)))).......((.------(((((........))))).))... ( -30.70) >sp_pn.0 341324 90 - 2038615/1648-1768 AAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUA---------------------AGGAA--CUGCGCAUUG-G------UCUUGUUUAGUCUUGAGAGGUCUU .(((((((((((((....))))))))).....))))((((((..(((((.---------------------..(((--(...((....-)------)...)))).))))))))))).... ( -31.00) >sp_mu.0 152460 98 - 2030921/1648-1768 AAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAAA---------------------AACGCA-AAGCACAUUAGGGGAACUUCUUGUUUAGUUUUGAGAGGGUAA ....((((((((((....)))))))))).....((((.((((....))))---------------------....((-(((((((..(((....)))..)))...)))))).)))).... ( -34.00) >consensus AAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUA_____________________AAGAUG_AGGAAUAAUGUGGACAUAUCGUAUUCAGUUUUGAAAGGUCAU ..((((((((((((....))))))))).....)))((((.....))))........................................................................ (-21.83 = -21.83 + -0.00)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:57:55 2006