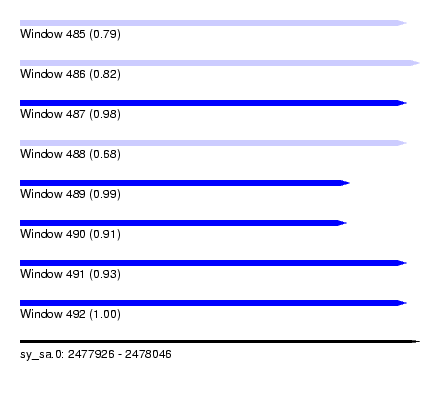
| Sequence ID | sy_sa.0 |
|---|---|
| Location | 2,477,926 – 2,478,046 |
| Length | 120 |
| Max. P | 0.995602 |

| Location | 2,477,926 – 2,478,042 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.96 |
| Mean single sequence MFE | -16.70 |
| Consensus MFE | -15.37 |
| Energy contribution | -13.60 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.33 |
| Mean z-score | 0.13 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 2477926 116 + 2516575/80-200 ----AAUUCGAAAUAAUAGAAACACAGUAACAAGGAAAGUAGAACUUACACUGCUGACACAGAGAGUCUUCGUAACGCUGAAAGAAGAUACAGAAAGAAAGUUUGAAAAUGGCCUUUGAG ----...........................((((..(...((((((.(.(((......))).(.((((((............)))))).).....).)))))).....)..)))).... ( -14.60) >sy_ha.0 2637323 114 + 2685015/80-200 ----AAAAAAACACAAAUUACACACUGUAAUAAGGAAAGUAGAACUUACACUGCUAAUACAGAGAGUCUCC--AAAGCUGAGAAGAGACAUUGAAAGAAAGUUUGAAAAUGGCCUUUGAG ----............(((((.....)))))((((..(...((((((.(.(((......)))...(((((.--...........))))).......).)))))).....)..)))).... ( -16.50) >sy_au.0 2370606 119 - 2809422/80-200 UAACAGUUUAAUGAAACGUAAACACAAUAAAGAGGAAAGUAAAACACACCCUGCUUAUACAGAGAGUCUUUA-GUAGCUGAGAGAAGAUUUUGAAAGCGUGUUUGAAAAUGGCCUUGGAG .....(((((........)))))........((((..(...((((((...(((......)))(((((((((.-..........)))))))))......)))))).....)..)))).... ( -19.00) >consensus ____AAUUAAAAAAAAAAUAAACACAGUAAAAAGGAAAGUAGAACUUACACUGCUAAUACAGAGAGUCUUC__AAAGCUGAGAGAAGAUAUUGAAAGAAAGUUUGAAAAUGGCCUUUGAG ...............................((((..(...((((((.(.(((......)))...((((((............)))))).......).)))))).....)..)))).... (-15.37 = -13.60 + -1.77)


| Location | 2,477,926 – 2,478,046 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.29 |
| Mean single sequence MFE | -29.67 |
| Consensus MFE | -28.60 |
| Energy contribution | -27.17 |
| Covariance contribution | -1.43 |
| Combinations/Pair | 1.21 |
| Mean z-score | -0.54 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 2477926 120 + 2516575/120-240 AGAACUUACACUGCUGACACAGAGAGUCUUCGUAACGCUGAAAGAAGAUACAGAAAGAAAGUUUGAAAAUGGCCUUUGAGUGUUGAUGCCAAUAUGAGGUAUCAACGGGCGCGCCCGUUA .((((((.(.(((......))).(.((((((............)))))).).....).)))))).....((((........(((((((((.......)))))))))(((....))))))) ( -29.60) >sy_ha.0 2637323 118 + 2685015/120-240 AGAACUUACACUGCUAAUACAGAGAGUCUCC--AAAGCUGAGAAGAGACAUUGAAAGAAAGUUUGAAAAUGGCCUUUGAGUGUUGAUGCCAAUACGAGGUGUCAACGGGUUCGCCCGUUA .((((((.(.(((......)))...(((((.--...........))))).......).)))))).......(((((...((((((....)))))))))))...((((((....)))))). ( -29.70) >sy_au.0 2370606 119 - 2809422/120-240 AAAACACACCCUGCUUAUACAGAGAGUCUUUA-GUAGCUGAGAGAAGAUUUUGAAAGCGUGUUUGAAAAUGGCCUUGGAGUGUUGAUGCCAAUAUGAGGUGUCUACGGGUUCGCCCGUUA ..(((((.((..(((....((((.((..((((-(...((......))...)))))..).).)))).....)))...)).)))))((((((.......)))))).(((((....))))).. ( -29.70) >consensus AGAACUUACACUGCUAAUACAGAGAGUCUUC__AAAGCUGAGAGAAGAUAUUGAAAGAAAGUUUGAAAAUGGCCUUUGAGUGUUGAUGCCAAUAUGAGGUGUCAACGGGUUCGCCCGUUA .((((((.(.(((......)))...((((((............)))))).......).)))))).......(((((...((((((....)))))))))))...((((((....)))))). (-28.60 = -27.17 + -1.43)

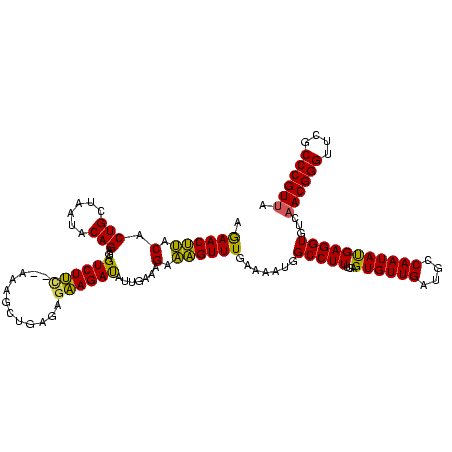
| Location | 2,477,926 – 2,478,042 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.07 |
| Mean single sequence MFE | -33.87 |
| Consensus MFE | -27.53 |
| Energy contribution | -27.77 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.92 |
| SVM RNA-class probability | 0.982775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 2477926 116 + 2516575/200-320 UGUUGAUGCCAAUAUGAGGUAUCAACGGGCGCGCCCGUUAUAGCGCUACAGUAUUAACAUUU--UGUUAAAUGGCGUACUGGAUUAUUCCACACGUUAUC--UUCUCGUUAAUACUCAUU .(((((((((.......))))))))).(((((..........)))))..(((((((((....--.(....(((((((..((((....)))).))))))).--..)..))))))))).... ( -35.90) >sy_ha.0 2637323 118 + 2685015/200-320 UGUUGAUGCCAAUACGAGGUGUCAACGGGUUCGCCCGUUAUAGCGAUACAGUAUUAACAUUUAUUGUUUAAUGGCGUACUGGA--AUUCUAGACGUCGCUAAUCAUCGUUAAUACUCAUU .(((((((((.......)))))))))(((....)))((..(((((((((((((........))))))(((.((((((.((((.--...)))))))))).)))..)))))))..))..... ( -38.00) >sy_au.0 2370606 115 - 2809422/200-320 UGUUGAUGCCAAUAUGAGGUGUCUACGGGUUCGCCCGUUAUAGCGAUACAGUAUUAACAUUGA-UGUUAAAUGGCGUACUGGA---UUCUUUACG-CACGAUUUUUUGUUAAUAAGUAUG .((((((((...(((..(.(((..(((((....))))).))).).)))..))))))))(((((-((..((((((((((..(..---..)..))))-).).))))..)))))))....... ( -27.70) >consensus UGUUGAUGCCAAUAUGAGGUGUCAACGGGUUCGCCCGUUAUAGCGAUACAGUAUUAACAUUUA_UGUUAAAUGGCGUACUGGA__AUUCUACACGUCACCA_UUAUCGUUAAUACUCAUU .(((((((((.......)))))))))(((....)))....((((((.......((((((.....)))))).((((((...(((....)))..)))))).......))))))......... (-27.53 = -27.77 + 0.23)

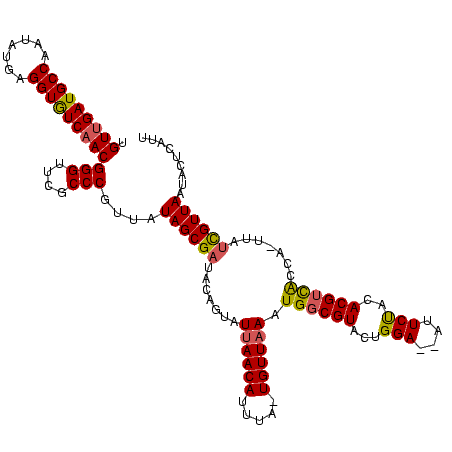
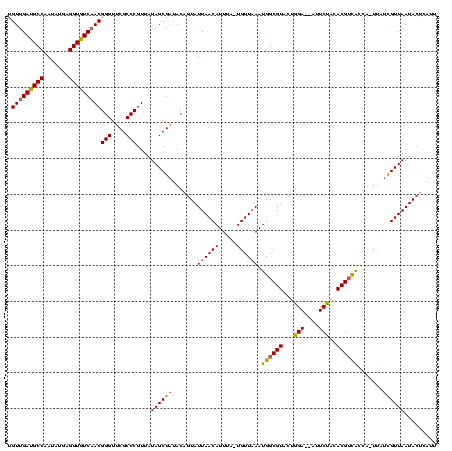
| Location | 2,477,926 – 2,478,042 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.07 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -19.19 |
| Energy contribution | -20.42 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 2477926 116 + 2516575/200-320 AAUGAGUAUUAACGAGAA--GAUAACGUGUGGAAUAAUCCAGUACGCCAUUUAACA--AAAUGUUAAUACUGUAGCGCUAUAACGGGCGCGCCCGUUGAUACCUCAUAUUGGCAUCAACA ....((((((((((...(--(((..((((((((....)))).))))..))))....--...))))))))))...(((((......)))))....((((((.((.......)).)))))). ( -32.30) >sy_ha.0 2637323 118 + 2685015/200-320 AAUGAGUAUUAACGAUGAUUAGCGACGUCUAGAAU--UCCAGUACGCCAUUAAACAAUAAAUGUUAAUACUGUAUCGCUAUAACGGGCGAACCCGUUGACACCUCGUAUUGGCAUCAACA ..(((((....((((.(..(((((((.....)...--..(((((...((((........))))....)))))..))))))(((((((....)))))))...).))))....)).)))... ( -25.40) >sy_au.0 2370606 115 - 2809422/200-320 CAUACUUAUUAACAAAAAAUCGUG-CGUAAAGAA---UCCAGUACGCCAUUUAACA-UCAAUGUUAAUACUGUAUCGCUAUAACGGGCGAACCCGUAGACACCUCAUAUUGGCAUCAACA .((((.((((((((.........(-((((..(..---..)..))))).........-....))))))))..)))).((((..(((((....))))).((....))....))))....... ( -20.85) >consensus AAUGAGUAUUAACGAAAA_UAGUGACGUAUAGAAU__UCCAGUACGCCAUUUAACA_UAAAUGUUAAUACUGUAUCGCUAUAACGGGCGAACCCGUUGACACCUCAUAUUGGCAUCAACA ....((((((((((......((((.((((..((....))...)))).))))..........))))))))))...(((((......)))))....(((((..((.......))..))))). (-19.19 = -20.42 + 1.23) # Strand winner: forward (1.00)

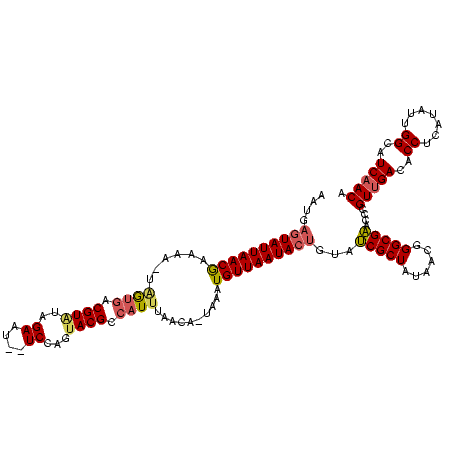
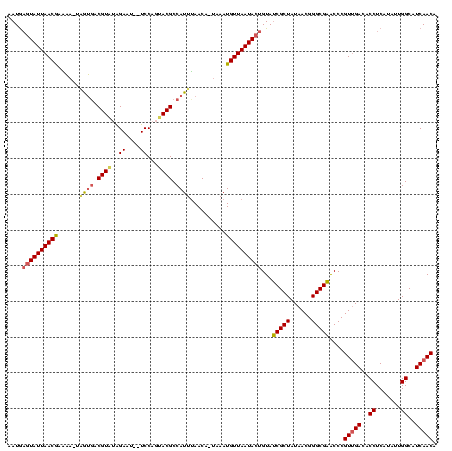
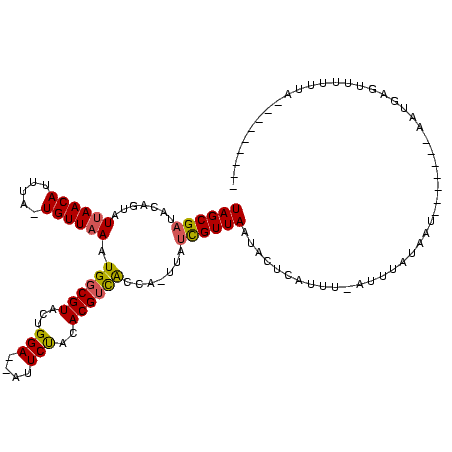
| Location | 2,477,926 – 2,478,025 |
|---|---|
| Length | 99 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 63.93 |
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -10.95 |
| Energy contribution | -11.07 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.45 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 2477926 99 + 2516575/240-360 UAGCGCUACAGUAUUAACAUUU--UGUUAAAUGGCGUACUGGAUUAUUCCACACGUUAUC--UUCUCGUUAAUACUCAUUUUAUUUAUAG--------AAUGGGUUUUUUA--------- (((((........(((((....--.)))))(((((((..((((....)))).))))))).--....)))))..((((((((((....)))--------)))))))......--------- ( -21.90) >sy_ha.0 2637323 117 + 2685015/240-360 UAGCGAUACAGUAUUAACAUUUAUUGUUUAAUGGCGUACUGGA--AUUCUAGACGUCGCUAAUCAUCGUUAAUACUCAUUU-ACUUAUAAUCAAGUUAAAUGAGUUUUUUAAUGUGAACC ((((...((((((........)))))).....(((((.((((.--...))))))))))))).(((.((((((.((((((((-((((......))).)))))))))...)))))))))... ( -34.00) >sy_au.0 2370606 99 - 2809422/240-360 UAGCGAUACAGUAUUAACAUUGA-UGUUAAAUGGCGUACUGGA---UUCUUUACG-CACGAUUUUUUGUUAAUAAGUAUGGGAUAGCACAU-------UACUAUAUCCUUA--------- .....((((..((((((((..((-.........(((((..(..---..)..))))-).....))..)))))))).))))((((((......-------.....))))))..--------- ( -17.84) >consensus UAGCGAUACAGUAUUAACAUUUA_UGUUAAAUGGCGUACUGGA__AUUCUACACGUCACCA_UUAUCGUUAAUACUCAUUU_AUUUAUAAU_______AAUGAGUUUUUUA_________ ((((((.......((((((.....)))))).((((((...(((....)))..)))))).......))))))................................................. (-10.95 = -11.07 + 0.12)


| Location | 2,477,926 – 2,478,024 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 57.10 |
| Mean single sequence MFE | -21.58 |
| Consensus MFE | -9.39 |
| Energy contribution | -10.51 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.43 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.910451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 2477926 98 + 2516575/360-480 -----UUUACGAAAAAUG--AUAACACGAUUCAGUCAACGU------------UACAUAUACA---ACAAUUUAGCGUACUGGAUAAGGUUACUUUAGUGAUAAUGUAACAAACAAAGGU -----.............--........(((((((..((((------------((.((.....---...)).)))))))))))))...(((((.(((....))).))))).......... ( -16.80) >sy_ha.0 2637323 120 + 2685015/360-480 UUUCAUUCGCCUAAAGUGUCACAUAACGAUUAAAUUAGCGUCCACUAUCUCAGUACACUUUUAUUCACAAAAUGGCGUACUGUUUGAGGUUACUUUAGCAAUAAAGUAAUAAAUAAAGGU .......(((.......(((.......))).......))).((.((....((((((.((.((........)).)).))))))....))(((((((((....))))))))).......)). ( -22.14) >sy_au.0 2370606 97 - 2809422/360-480 -CUUACUGACUUUAAUUGUGAUA--AUUGUUCAGUAAGCAU-----------AUUUACUUUUA---------AUGCGUACUGAAUAAGGUUAUUUCAGCGAUGGAAUAACAAAUAAAGGU -........(((((.........--.((((((((((.((((-----------...........---------)))).)))))))))).(((((((((....)))))))))...))))).. ( -25.80) >consensus __U_AUUCACCUAAAAUGU_AUA__ACGAUUCAGUAAGCGU____________UACACUUUUA___ACAA__UAGCGUACUGAAUAAGGUUACUUUAGCGAUAAAGUAACAAAUAAAGGU ............................(((((((..((((.................................)))))))))))...(((((((((....))))))))).......... ( -9.39 = -10.51 + 1.12)

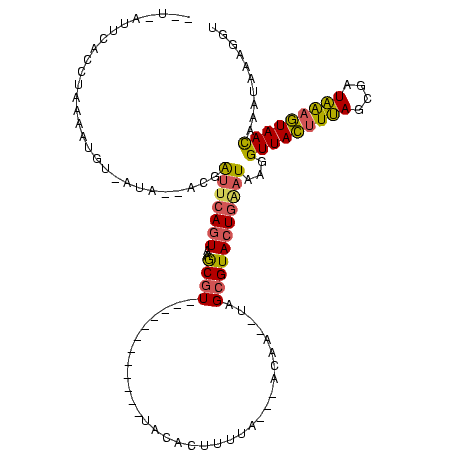
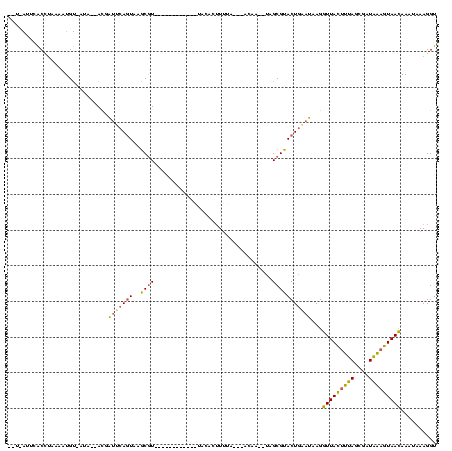
| Location | 2,477,926 – 2,478,042 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.68 |
| Mean single sequence MFE | -45.86 |
| Consensus MFE | -43.97 |
| Energy contribution | -43.77 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.21 |
| Mean z-score | -5.86 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 2477926 116 + 2516575/435-555 CGUACUGGAUAAGGUUACUUUAGUGAUAAUGUAACAAACAAAGGUGGUACCGCGAGC--UAAGCUUUCGUCCUUUACAUCCGAAU--UUUCGGGUUUAAAGGACGGGAGCUUUUUUUAUU ....((.......(((((.(((....))).)))))......))(((....)))(((.--.(((((..(((((((((.((((((..--..)))))).)))))))))..)))))..)))... ( -42.02) >sy_ha.0 2637323 118 + 2685015/435-555 CGUACUGUUUGAGGUUACUUUAGCAAUAAAGUAAUAAAUAAAGGUGGUACCGCGAAA--CAGGCUUUCGUCCUUUACAUCCGAUGCAUAUCGGGUUUAAAGGACGGGAGCUUUUUUUAUU .((((..(((...(((((((((....)))))))))......)))..))))...((((--.(((((..(((((((((.(((((((....))))))).)))))))))..))))).))))... ( -50.00) >sy_au.0 2370606 119 - 2809422/435-555 CGUACUGAAUAAGGUUAUUUCAGCGAUGGAAUAACAAAUAAAGGUGGUACCGCGAAACAUAAGCUUUCGUCCUUUUUAUCCGAUUCAU-UCGGGUACGAAGGACGGAAGCUUUUUUUAUU .((((((......(((((((((....))))))))).........))))))...((((...(((((((((((((((.(((((((.....-))))))).))))))))))))))).))))... ( -45.56) >consensus CGUACUGAAUAAGGUUACUUUAGCGAUAAAGUAACAAAUAAAGGUGGUACCGCGAAA__UAAGCUUUCGUCCUUUACAUCCGAUUCAU_UCGGGUUUAAAGGACGGGAGCUUUUUUUAUU .((((((..(...(((((((((....)))))))))......)..))))))..........((((((((((((((((.(((((((....))))))).))))))))))))))))........ (-43.97 = -43.77 + -0.21)

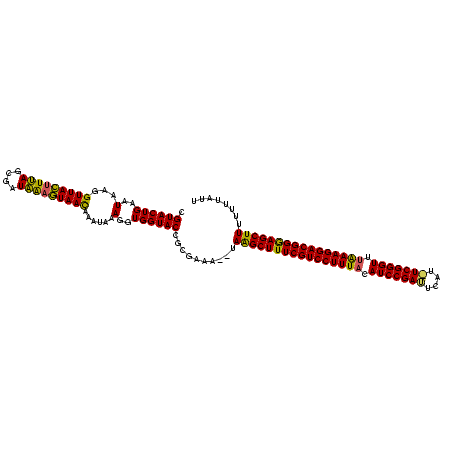
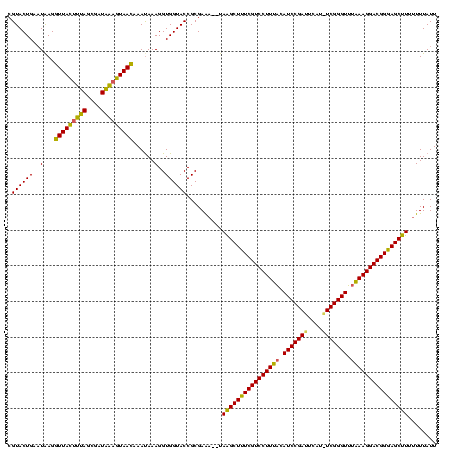
| Location | 2,477,926 – 2,478,042 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.68 |
| Mean single sequence MFE | -31.30 |
| Consensus MFE | -27.35 |
| Energy contribution | -28.58 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.55 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995602 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 2477926 116 + 2516575/435-555 AAUAAAAAAAGCUCCCGUCCUUUAAACCCGAAA--AUUCGGAUGUAAAGGACGAAAGCUUA--GCUCGCGGUACCACCUUUGUUUGUUACAUUAUCACUAAAGUAACCUUAUCCAGUACG ........(((((..(((((((((...((((..--..))))...)))))))))..))))).--.......((((...........(((((.(((....))).)))))........)))). ( -29.41) >sy_ha.0 2637323 118 + 2685015/435-555 AAUAAAAAAAGCUCCCGUCCUUUAAACCCGAUAUGCAUCGGAUGUAAAGGACGAAAGCCUG--UUUCGCGGUACCACCUUUAUUUAUUACUUUAUUGCUAAAGUAACCUCAAACAGUACG ..........(((..(((((((((...(((((....)))))...)))))))))..)))(((--(((...((.....))........((((((((....))))))))....)))))).... ( -33.00) >sy_au.0 2370606 119 - 2809422/435-555 AAUAAAAAAAGCUUCCGUCCUUCGUACCCGA-AUGAAUCGGAUAAAAAGGACGAAAGCUUAUGUUUCGCGGUACCACCUUUAUUUGUUAUUCCAUCGCUGAAAUAACCUUAUUCAGUACG ........((((((.(((((((..((.((((-.....)))).))..))))))).)))))).(((((((((((.....................))))).))))))............... ( -31.50) >consensus AAUAAAAAAAGCUCCCGUCCUUUAAACCCGA_AUGAAUCGGAUGUAAAGGACGAAAGCUUA__UUUCGCGGUACCACCUUUAUUUGUUACUUUAUCGCUAAAGUAACCUUAUACAGUACG ........(((((..(((((((((...(((((....)))))...)))))))))..)))))..........((((...........(((((((((....)))))))))........)))). (-27.35 = -28.58 + 1.23) # Strand winner: forward (1.00)

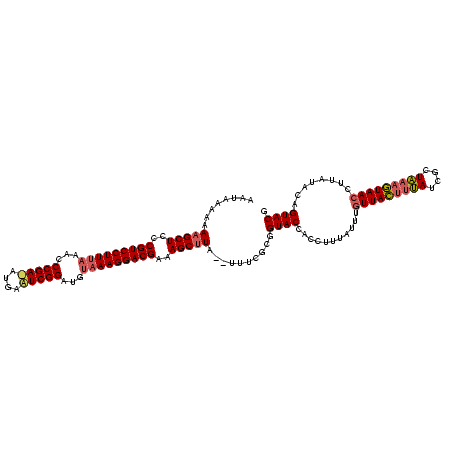
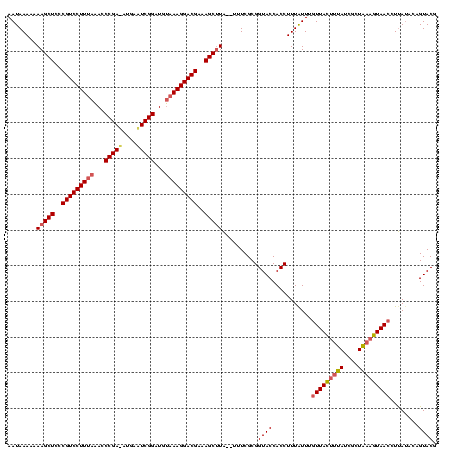
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:57:23 2006