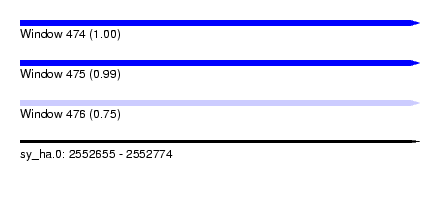
| Sequence ID | sy_ha.0 |
|---|---|
| Location | 2,552,655 – 2,552,774 |
| Length | 119 |
| Max. P | 0.999640 |

| Location | 2,552,655 – 2,552,774 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.16 |
| Mean single sequence MFE | -27.77 |
| Consensus MFE | -25.17 |
| Energy contribution | -24.73 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.91 |
| SVM decision value | 3.82 |
| SVM RNA-class probability | 0.999640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
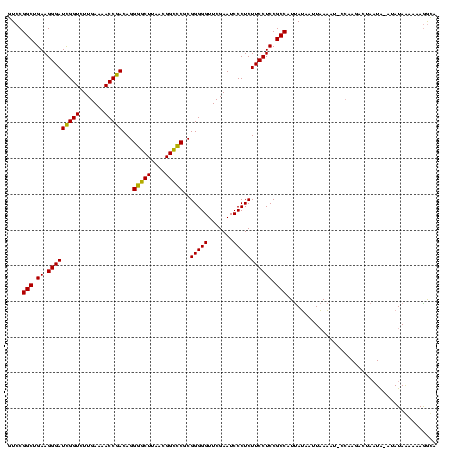
>sy_ha.0 2552655 119 + 2685015/0-120 UAAAAAAGACAAGUACUCAAUAUGUUAUAAAUUGAGUACUUGUCUUUUUUAUAUUUAUUAGUCUUGG-AUUUUAAUUAUAAUGGCGGAGGAAGAGGGAUUCGAACCCCCGCGGCCCGUUA (((((((((((((((((((((.........)))))))))))))))))))))................-..........(((((((.(.((....(((.......))))).).).)))))) ( -36.80) >sy_au.0 2289354 108 - 2809422/0-120 UAAAAAAGGUAAAUACUCAAAAU--------UUGAGCACUUACCUUCAUUAUAUAUAGAA---UUUA-AUAUAAAUAUUGAUGGCGGAGGAAGAGGGAUUCGAACCCCCGCGAGCCGUUA ......((((((...((((....--------.))))...))))))(((.(((.((((...---....-.)))).))).)))((((((.....(.(((........)))).....)))))) ( -23.00) >sy_sa.0 2363694 116 + 2516575/0-120 UUAAAAAAGCAAGCAUCCAAAUUUUUC---AUUGAAUACUUGCUUUAUCUAUAU-CAUUAGUCUUGGUAUUUUAAAUAUAAUGGCGGAGGAAGAGGGAUUCGAACCCCCGCGGGCCGUUA (((((((((((((.((.(((.......---.))).)).))))))))....((((-((.......)))))))))))...(((((((...((....(((.......)))))....))))))) ( -23.50) >consensus UAAAAAAGGCAAGUACUCAAAAU_UU____AUUGAGUACUUGCCUUAUUUAUAU_UAUUAGUCUUGG_AUUUUAAUUAUAAUGGCGGAGGAAGAGGGAUUCGAACCCCCGCGGGCCGUUA .....(((((((((((((((...........)))))))))))))))...................................((((((.....(.(((........)))).....)))))) (-25.17 = -24.73 + -0.44)

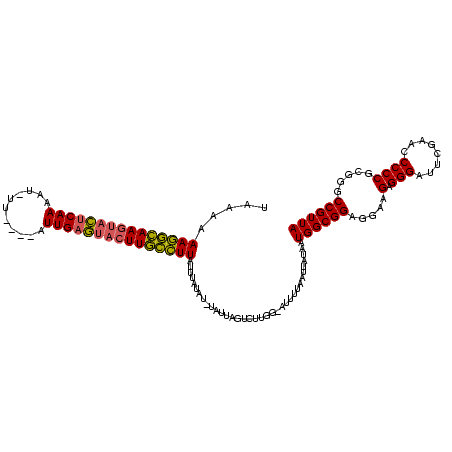
| Location | 2,552,655 – 2,552,774 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.16 |
| Mean single sequence MFE | -34.63 |
| Consensus MFE | -35.82 |
| Energy contribution | -33.83 |
| Covariance contribution | -1.98 |
| Combinations/Pair | 1.34 |
| Mean z-score | -5.16 |
| Structure conservation index | 1.03 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
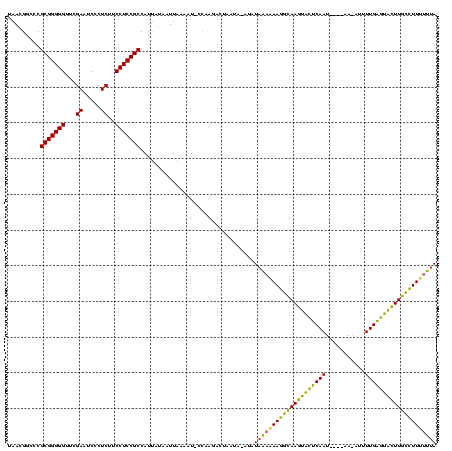
>sy_ha.0 2552655 119 + 2685015/0-120 UAACGGGCCGCGGGGGUUCGAAUCCCUCUUCCUCCGCCAUUAUAAUUAAAAU-CCAAGACUAAUAAAUAUAAAAAAGACAAGUACUCAAUUUAUAACAUAUUGAGUACUUGUCUUUUUUA ....(((..(((((((...((.....))..)))))))..(((....)))..)-))..............(((((((((((((((((((((.........))))))))))))))))))))) ( -39.70) >sy_au.0 2289354 108 - 2809422/0-120 UAACGGCUCGCGGGGGUUCGAAUCCCUCUUCCUCCGCCAUCAAUAUUUAUAU-UAAA---UUCUAUAUAUAAUGAAGGUAAGUGCUCAA--------AUUUUGAGUAUUUACCUUUUUUA .........(((((((...((.....))..)))))))..........(((((-....---....)))))(((.(((((((((((((((.--------....))))))))))))))).))) ( -32.20) >sy_sa.0 2363694 116 + 2516575/0-120 UAACGGCCCGCGGGGGUUCGAAUCCCUCUUCCUCCGCCAUUAUAUUUAAAAUACCAAGACUAAUG-AUAUAGAUAAAGCAAGUAUUCAAU---GAAAAAUUUGGAUGCUUGCUUUUUUAA .........(((((((...((.....))..)))))))............................-...((((.(((((((((((((((.---.......))))))))))))))))))). ( -32.00) >consensus UAACGGCCCGCGGGGGUUCGAAUCCCUCUUCCUCCGCCAUUAUAAUUAAAAU_CCAAGACUAAUA_AUAUAAAAAAGGCAAGUACUCAAU____AA_AUUUUGAGUACUUGCCUUUUUUA .........(((((((...((.....))..)))))))................................((((((((((((((((((((...........)))))))))))))))))))) (-35.82 = -33.83 + -1.98) # Strand winner: reverse (1.00)

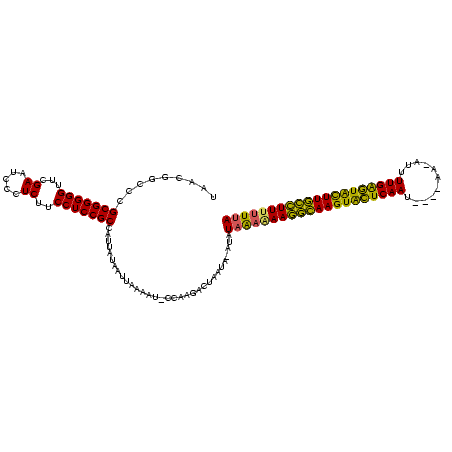
| Location | 2,552,655 – 2,552,774 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.40 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -30.31 |
| Energy contribution | -29.20 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
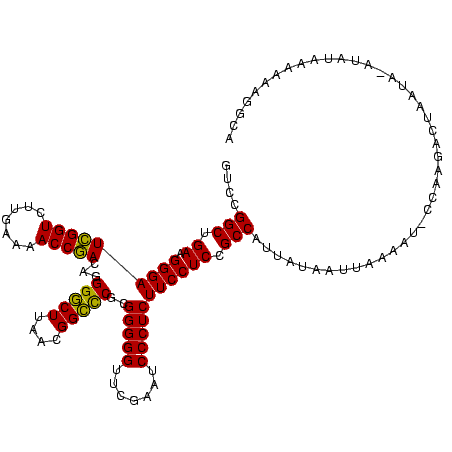
>sy_ha.0 2552655 119 + 2685015/40-160 GUCCGGCUGAAGGGAUCGGUCUUGAAAACCGACAGGGCCUUAACGGGCCGCGGGGGUUCGAAUCCCUCUUCCUCCGCCAUUAUAAUUAAAAU-CCAAGACUAAUAAAUAUAAAAAAGACA ((((........)))).(((((((...........(((((....)))))(((((((...((.....))..)))))))...............-.)))))))................... ( -32.90) >sy_au.0 2289354 116 - 2809422/40-160 GUCCGGCUGAAGGGAUCGGUCUUGAAAACCGACAGGGGCUUAACGGCUCGCGGGGGUUCGAAUCCCUCUUCCUCCGCCAUCAAUAUUUAUAU-UAAA---UUCUAUAUAUAAUGAAGGUA ..(((((.((.(((((((((.......)))))...(((((....)))))..(((((.......))))))))))).))).(((.(((.((((.-....---...)))).))).))).)).. ( -31.80) >sy_sa.0 2363694 119 + 2516575/40-160 GUCCGGCUGAAGGGAUUGGUCUUGAAAACCAACAGGGGCUUAACGGCCCGCGGGGGUUCGAAUCCCUCUUCCUCCGCCAUUAUAUUUAAAAUACCAAGACUAAUG-AUAUAGAUAAAGCA .(((.......)))((((((((((...........(((((....)))))(((((((...((.....))..))))))).................)))))))))).-.............. ( -34.90) >consensus GUCCGGCUGAAGGGAUCGGUCUUGAAAACCGACAGGGGCUUAACGGCCCGCGGGGGUUCGAAUCCCUCUUCCUCCGCCAUUAUAAUUAAAAU_CCAAGACUAAUA_AUAUAAAAAAGGCA ....(((.((.(((((((((.......)))))...(((((....)))))..(((((.......))))))))))).))).......................................... (-30.31 = -29.20 + -1.11) # Strand winner: reverse (0.69)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:57:10 2006