| Sequence ID | sy_ha.0 |
|---|---|
| Location | 2,495,471 – 2,495,591 |
| Length | 120 |
| Max. P | 1.000000 |

| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.79 |
| Mean single sequence MFE | -27.47 |
| Consensus MFE | -24.28 |
| Energy contribution | -23.07 |
| Covariance contribution | -1.21 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 120 + 2685015/0-120 CAAAGAUGAUGAGGUUAAUAGGUUCGAGGUGGAAGCGUGGUGACACGUGGAGCUGACGAAUACUAAUCGAUUGAAGACUUAAUCAAUUUAUUUCAAUGUUUUGCGAAGCAAAAUGAUUUA ....((.((((((((((....(((((.(((....(((((....)))))...)))..)))))..)))))((((((....))))))...)))))))...(((((((...)))))))...... ( -30.30) >sy_au.0 2231801 116 - 2809422/0-120 CAAAGAUGAUGAGGUUAAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUCGAUCGAAGACUUAAUCAAAAUA----AAUGUUUUGCGAAGCAAAAUCACUUU .((((.((((.(((((..((((((((.(((....(((((....)))))...)))..))))).))).((....)).))))).)))).....----...(((((((...)))))))..)))) ( -27.70) >sy_sa.0 2306606 118 + 2516575/0-120 CAAAGAUGAUGAGGUUAAUAGGUUCGAGGUGGAAGCAUAGCGAUAUGUGCAGCUGACGAAUACUAAUCGAUCGAAGACUUAAUCAAAUU--UUAAAUGUUUUGUUUGGUUGAAUCAUAUU ......((((..(((((....(((((.(((....(((((......))))).)))..)))))..))))).))))..((.((((((((((.--...........)))))))))).))..... ( -24.40) >consensus CAAAGAUGAUGAGGUUAAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUCGAUCGAAGACUUAAUCAAAUUA_UU_AAUGUUUUGCGAAGCAAAAUCAUUUU ......((((.(((((..((((((((.(((....(((((....)))))...)))..))))).))).((....)).))))).))))............(((((((...)))))))...... (-24.28 = -23.07 + -1.21) # Strand winner: reverse (1.00)

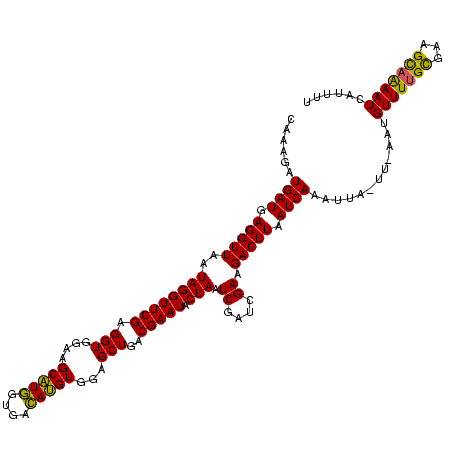

| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -54.40 |
| Consensus MFE | -54.40 |
| Energy contribution | -54.40 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.58 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 120 + 2685015/320-440 UUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUC ......(((..((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..)).(((((((....))).))))))). ( -54.40) >sy_au.0 2231801 120 - 2809422/320-440 UUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUC ......(((..((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..)).(((((((....))).))))))). ( -54.40) >sy_sa.0 2306606 120 + 2516575/320-440 UUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUC ......(((..((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..)).(((((((....))).))))))). ( -54.40) >consensus UUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUC ......(((..((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..)).(((((((....))).))))))). (-54.40 = -54.40 + -0.00) # Strand winner: reverse (1.00)

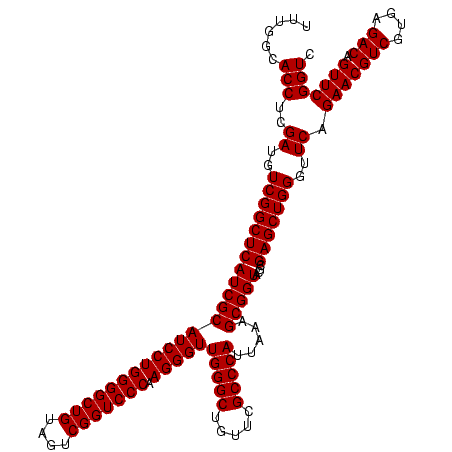
| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -50.30 |
| Consensus MFE | -50.30 |
| Energy contribution | -50.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.847189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 120 + 2685015/360-480 ACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGG ....(((.((((((((...((.......))...))))))))..))).((..((((......))))((((((((((((((....))))))).)))))(((((.....))))).....)))) ( -50.30) >sy_au.0 2231801 120 - 2809422/360-480 ACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGG ....(((.((((((((...((.......))...))))))))..))).((..((((......))))((((((((((((((....))))))).)))))(((((.....))))).....)))) ( -50.30) >sy_sa.0 2306606 120 + 2516575/360-480 ACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGG ....(((.((((((((...((.......))...))))))))..))).((..((((......))))((((((((((((((....))))))).)))))(((((.....))))).....)))) ( -50.30) >consensus ACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGG ....(((.((((((((...((.......))...))))))))..))).((..((((......))))((((((((((((((....))))))).)))))(((((.....))))).....)))) (-50.30 = -50.30 + 0.00) # Strand winner: reverse (0.98)

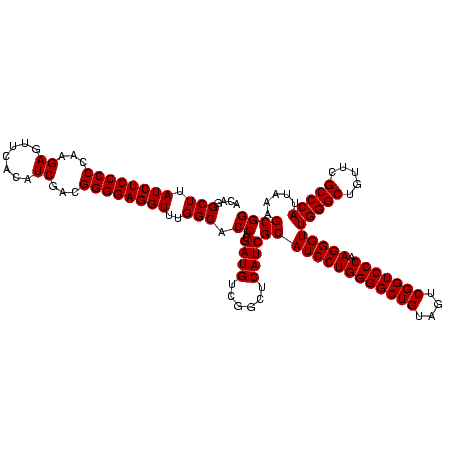
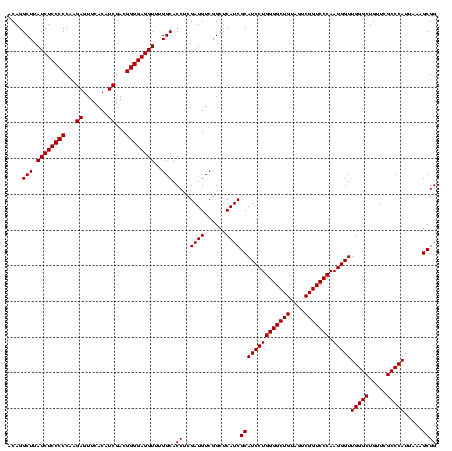
| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -34.36 |
| Energy contribution | -33.70 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.88 |
| Structure conservation index | 1.02 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 120 + 2685015/560-680 CUGCUCGACUUGUAGGUCUCGCAGUCAAGCUCCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCC ((((..((((....))))..))))(((((.(((((((((......))......(((((.........))))))))))))...))))).(((((((...........)))))))....... ( -34.60) >sy_au.0 2231801 120 - 2809422/560-680 CUGCUCGACUUGUAGGUCUCGCAGUCAAGCUCCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCC ((((..((((....))))..))))(((((.(((((((((......))......(((((.........))))))))))))...))))).(((((((...........)))))))....... ( -34.60) >sy_sa.0 2306606 120 + 2516575/560-680 CUGCUCGACUUGUAAGUCUCGCAGUCAAGCUUCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGAAACCUUUGAGCGCCUCCGUUACUCUUUAGGAGGCGACCGCCC ((((..((((....))))..))))(((((.(((((((((......))......(((((.........))))))))))))...))))).(((((((...........)))))))....... ( -31.90) >consensus CUGCUCGACUUGUAGGUCUCGCAGUCAAGCUCCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCC ((((..((((....))))..))))(((((.(((((((((......))......(((((.........))))))))))))...))))).(((((((...........)))))))....... (-34.36 = -33.70 + -0.66)

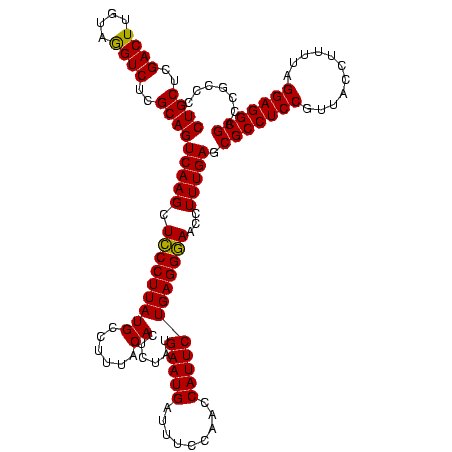
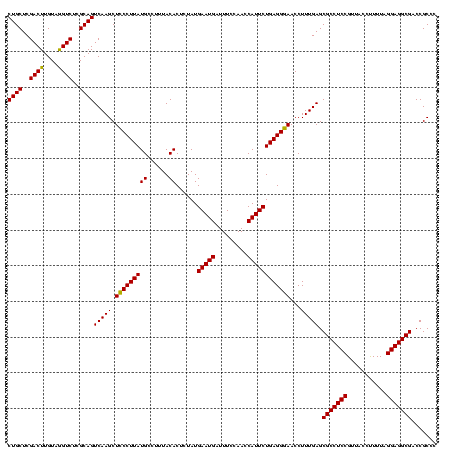
| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -43.50 |
| Consensus MFE | -43.78 |
| Energy contribution | -43.33 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.04 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.993672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
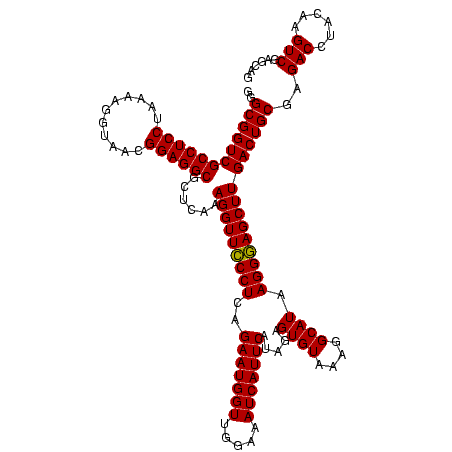
>sy_ha.0 2495471 120 + 2685015/560-680 GGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAG ..((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((((....)))).)))))))))))))))..(((......)))...... ( -43.90) >sy_au.0 2231801 120 - 2809422/560-680 GGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAG ..((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((((....)))).)))))))))))))))..(((......)))...... ( -43.90) >sy_sa.0 2306606 120 + 2516575/560-680 GGGCGGUCGCCUCCUAAAGAGUAACGGAGGCGCUCAAAGGUUUCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGAAGCUUGACUGCGAGACUUACAAGUCGAGCAG ..((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((((....)))).)))))))))))))))..((((....))))...... ( -42.70) >consensus GGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAG ..((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((((....)))).)))))))))))))))..(((......)))...... (-43.78 = -43.33 + -0.44) # Strand winner: reverse (1.00)

| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -42.77 |
| Consensus MFE | -41.11 |
| Energy contribution | -40.67 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
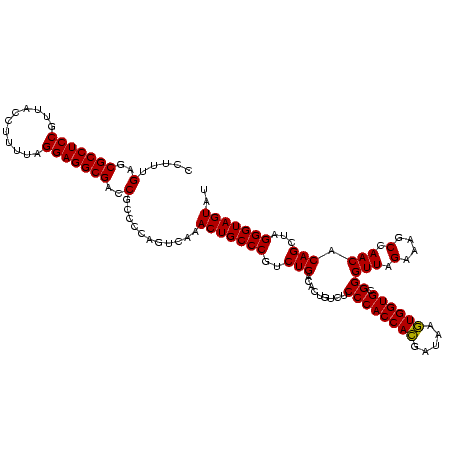
>sy_ha.0 2495471 120 + 2685015/640-760 CCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAGCUAGGGUAGUAU .....(..(((((((...........)))))))..)...........(((((((.((((((.(((.....((((((.....))))))))))))))).(((.......))).))))))).. ( -44.70) >sy_au.0 2231801 120 - 2809422/640-760 CCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAGCUAGGGUAGUAU .....(..(((((((...........)))))))..)...........(((((((..(((((.(((.....((((((.....))))))))))))))..(((.......))).))))))).. ( -43.20) >sy_sa.0 2306606 120 + 2516575/640-760 CCUUUGAGCGCCUCCGUUACUCUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCAUCUGACACUGUCUCCCACCAUGAUAAAUGGUGCGGGUUAGAAAUCCAACACAGCGAGGGUAGUAU .....(..(((((((...........)))))))..)...........(((((((.((((((.(((.....((((((.....)))))))))))))))..(((......).))))))))).. ( -40.40) >consensus CCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAGCUAGGGUAGUAU .....(..(((((((...........)))))))..)...........(((((((..(((.........((((((((.....)))))).))(((.(....).))).)))...))))))).. (-41.11 = -40.67 + -0.44)


| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -47.40 |
| Consensus MFE | -47.85 |
| Energy contribution | -47.30 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.06 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953615 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
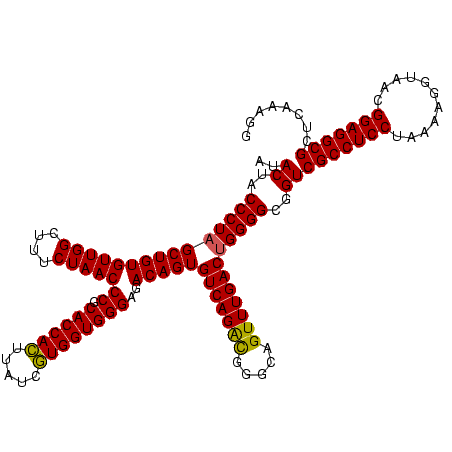
>sy_ha.0 2495471 120 + 2685015/640-760 AUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGG ..((..(((((((((((((((....)))))((.((((((.....))))))))..)))))(((((((.....))))))))))))..))(((((((...........)))))))........ ( -49.10) >sy_au.0 2231801 120 - 2809422/640-760 AUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGGCGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGG ..((..(((((((((((((((....)))))((.((((((.....))))))))..)))))(((((((.....))))))))))))..))(((((((...........)))))))........ ( -48.60) >sy_sa.0 2306606 120 + 2516575/640-760 AUACUACCCUCGCUGUGUUGGAUUUCUAACCCGCACCAUUUAUCAUGGUGGGAGACAGUGUCAGAUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGAGUAACGGAGGCGCUCAAAGG ..((..((((.((((((((((....)))))((.((((((.....))))))))..)))))(((((((.....))))))).))))..))(((((((...........)))))))........ ( -44.50) >consensus AUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGG ..((..(((((((((((((((....)))))((.((((((.....))))))))..)))))(((((((.....))))))))))))..))(((((((...........)))))))........ (-47.85 = -47.30 + -0.55) # Strand winner: reverse (1.00)

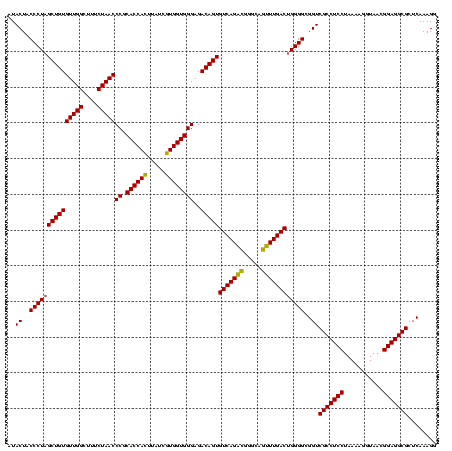
| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -33.21 |
| Energy contribution | -32.77 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 120 + 2685015/680-800 CAGUCAAACUGCCCGUCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAGCGCUCACGUUUCAA .......(((((((.((((((.(((.....((((((.....))))))))))))))).(((.......))).)))))))........(((((.....(....)...))))).......... ( -36.80) >sy_au.0 2231801 120 - 2809422/680-800 CAGUCAAACUGCCCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAGCGCUCACGUUUCAA .......(((((((..(((((.(((.....((((((.....))))))))))))))..(((.......))).)))))))........(((((.....(....)...))))).......... ( -35.30) >sy_sa.0 2306606 120 + 2516575/680-800 CAGUCAAACUGCCCAUCUGACACUGUCUCCCACCAUGAUAAAUGGUGCGGGUUAGAAAUCCAACACAGCGAGGGUAGUAUCCCACCAACGCCUCCACGUAAGCUAGCGCUCACGUAUCUC .......(((((((.((((((.(((.....((((((.....)))))))))))))))..(((......).))))))))).................((((.(((....))).))))..... ( -32.90) >consensus CAGUCAAACUGCCCGUCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAAAGCCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAGCGCUCACGUUUCAA .......(((((((..(((.........((((((((.....)))))).))(((.(....).))).)))...))))))).................((((.(((....))).))))..... (-33.21 = -32.77 + -0.44)

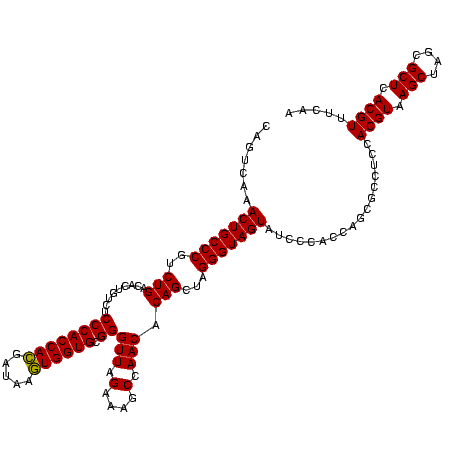
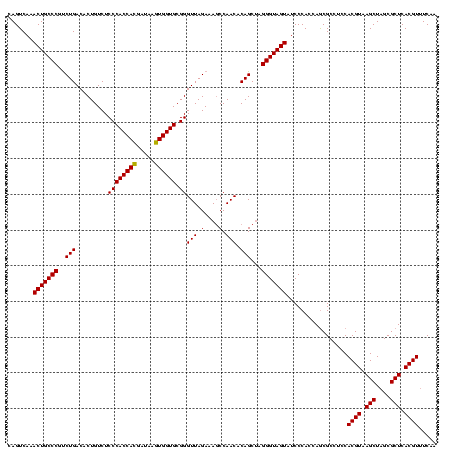
| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -46.37 |
| Consensus MFE | -45.38 |
| Energy contribution | -44.50 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 120 + 2685015/680-800 UUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGACGGGCAGUUUGACUG .....((((((((....)))))))).((.(((((((.((.......))))))).)).)).(((.(((...((.((((((.....))))))))))).)))(((((((.....))))))).. ( -46.40) >sy_au.0 2231801 120 - 2809422/680-800 UUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGGCGGGCAGUUUGACUG .....((((((((....)))))))).((.(((((((.((.......))))))).)).)).(((.(((...((.((((((.....))))))))))).)))(((((((.....))))))).. ( -45.90) >sy_sa.0 2306606 120 + 2516575/680-800 GAGAUACGUGAGCGCUAGCUUACGUGGAGGCGUUGGUGGGAUACUACCCUCGCUGUGUUGGAUUUCUAACCCGCACCAUUUAUCAUGGUGGGAGACAGUGUCAGAUGGGCAGUUUGACUG (((.(((((((((....)))))))))........(((((....))))))))((((((((((....)))))((.((((((.....))))))))..)))))(((((((.....))))))).. ( -46.80) >consensus UUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACUUAUCGUGGUGGGAGACAGUGUCAGACGGGCAGUUUGACUG .....((((((((....)))))))).........(((((....)))))...((((((((((....)))))((.((((((.....))))))))..)))))(((((((.....))))))).. (-45.38 = -44.50 + -0.88) # Strand winner: reverse (1.00)

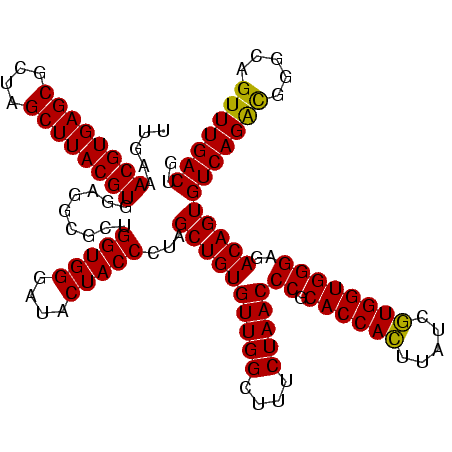
| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -46.90 |
| Consensus MFE | -43.63 |
| Energy contribution | -43.97 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 120 + 2685015/720-840 UUGAAAUUCGGCACAGCUUGUACAGGAUAGGUAGGAGCCUUUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACU ..((((..((((((((((.(.............((.(((((....((((((((....)))))))).))))).))(((((....)))))).))))))))))..)))).............. ( -45.50) >sy_au.0 2231801 120 - 2809422/720-840 UUGAAAUUCGGCACAGCUUGUACAGGAUAGGUAGGAGCCUUUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACU ..((((..((((((((((.(.............((.(((((....((((((((....)))))))).))))).))(((((....)))))).))))))))))..)))).............. ( -45.50) >sy_sa.0 2306606 120 + 2516575/720-840 UUGAAAUUCGGCACAGCUUGUACAGGAUAGGUAGGAGCCUGAGAUACGUGAGCGCUAGCUUACGUGGAGGCGUUGGUGGGAUACUACCCUCGCUGUGUUGGAUUUCUAACCCGCACCAUU ..(((((((((((((((((.(((.......))).))((((....(((((((((....))))))))).))))(..(((((....)))))..)))))))))))))))).............. ( -49.70) >consensus UUGAAAUUCGGCACAGCUUGUACAGGAUAGGUAGGAGCCUUUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCUUUCUAACCCGCACCACU ..((((.((((((((((((.(((.......))).))(((((....((((((((....)))))))).)))))...(((((....)))))...)))))))))).)))).............. (-43.63 = -43.97 + 0.33) # Strand winner: reverse (1.00)

| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -36.83 |
| Consensus MFE | -36.92 |
| Energy contribution | -36.70 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.47 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 120 + 2685015/1120-1240 GCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUUUAGGGUGCAAGCCCAGAAGAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAUCAAAAACACAGGUCUCUGCUAAACCGUAA ........(((((....(....).....)))))(((((((.((((....)))).))))))).((((.((....((....))..((((...........)))).)).)))).......... ( -37.30) >sy_au.0 2231801 120 - 2809422/1120-1240 GCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUUUAGGGUUAACGCCCAGAAGAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAUCAAAAACACAGGUCUCUGCUAAACCGUAA ........(((((....(....).....)))))(((((((.((((....)))).))))))).((((.((....((....))..((((...........)))).)).)))).......... ( -36.60) >sy_sa.0 2306606 120 + 2516575/1120-1240 GCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUUUAGGGUUAACGCCCAGGAGAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAUCAAAAACACAGGUCUCUGCUAAACCGUAA ........(((((....(....).....)))))((((((..((((....))))..)))))).((((.((....((....))..((((...........)))).)).)))).......... ( -36.60) >consensus GCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUUUAGGGUUAACGCCCAGAAGAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAUCAAAAACACAGGUCUCUGCUAAACCGUAA ........(((((....(....).....)))))(((((((.((((....)))).))))))).((((.((....((....))..((((...........)))).)).)))).......... (-36.92 = -36.70 + -0.22) # Strand winner: reverse (0.65)

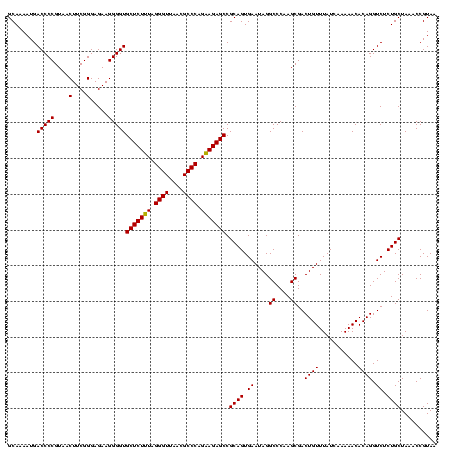
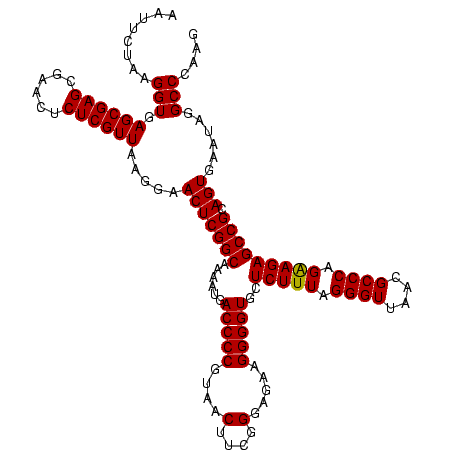
| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -39.33 |
| Consensus MFE | -39.42 |
| Energy contribution | -39.20 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.36 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.552209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 120 + 2685015/1160-1280 AAUUCUAAGGUGAGCGAGCGAACUCUCGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUUUAGGGUGCAAGCCCAGAAGAGCCGCAGUGAAUAGGCCCAAG ........(((.((((((......)))))).....(((((((......(((((....(....).....)))))..(((((.((((....)))).))))))))).)))......))).... ( -39.80) >sy_au.0 2231801 120 - 2809422/1160-1280 AAUUCUAAGGUGAGCGAGCGAACUCUCGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUUUAGGGUUAACGCCCAGAAGAGCCGCAGUGAAUAGGCCCAAG ........(((.((((((......)))))).....(((((((......(((((....(....).....)))))..(((((.((((....)))).))))))))).)))......))).... ( -39.10) >sy_sa.0 2306606 120 + 2516575/1160-1280 AAUUCUAAGGUGAGCGAGCGAACUCUCGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUUUAGGGUUAACGCCCAGGAGAGCCGCAGUGAAUAGGCCCAAG ........(((.((((((......)))))).....(((((((......(((((....(....).....)))))..((((..((((....))))..)))))))).)))......))).... ( -39.10) >consensus AAUUCUAAGGUGAGCGAGCGAACUCUCGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUUUAGGGUUAACGCCCAGAAGAGCCGCAGUGAAUAGGCCCAAG ........(((.((((((......)))))).....(((((((......(((((....(....).....)))))..(((((.((((....)))).))))))))).)))......))).... (-39.42 = -39.20 + -0.22) # Strand winner: reverse (0.98)

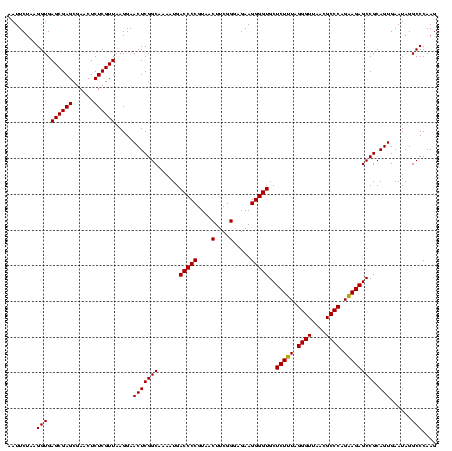
| Location | 2,495,471 – 2,495,590 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.42 |
| Mean single sequence MFE | -28.37 |
| Consensus MFE | -26.31 |
| Energy contribution | -25.53 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943568 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 119 + 2685015/1320-1440 UAUUAUCUAUCUAGAGGCUUUUCUCGGCAGUGUGAAAUCAACGACUCGAGGAAACAAUUUCCUCUCCCCAUCACAGCUCAAUCUUAA-GAGUGCCGGAUUUGCCUAACACUCAAUCUCAC ..............((((...((.((((..(((((..((...))...((((((.....))))))......)))))((((........-))))))))))...))))............... ( -29.20) >sy_au.0 2231801 120 - 2809422/1320-1440 UAUUUUCUAUCUAGAGGCUUUUCUCGGCAGUGUGAAAUCAACGACUCGAAGACACAAUGUCUUCUCCCCAUCACAGCUCAGCCUUAACGAGUACCGGAUUUGCCUAAUACUCAGCCUUAC .............((((((......((((((((((..((...))...((((((.....))))))......))))).))..))).....(((((..((.....))...))))))))))).. ( -28.90) >sy_sa.0 2306606 119 + 2516575/1320-1440 ACAAUUCUAGCUAGAGGCUUUUCUUGGCAGUGUGAAAUCAACGACUCGAGGAAACAAUUUCCUCUCCCCAUCACAGCUCAAUCUUAA-GAGUGCCGGAUUUGCCUAACACUCAAUCUCAC ..............((((...(((.(((..(((((..((...))...((((((.....))))))......)))))((((........-))))))))))...))))............... ( -27.00) >consensus UAUUUUCUAUCUAGAGGCUUUUCUCGGCAGUGUGAAAUCAACGACUCGAGGAAACAAUUUCCUCUCCCCAUCACAGCUCAAUCUUAA_GAGUGCCGGAUUUGCCUAACACUCAAUCUCAC ..............((((...((.((((..(((((..((...))...((((((.....))))))......)))))((((.........))))))))))...))))............... (-26.31 = -25.53 + -0.77)

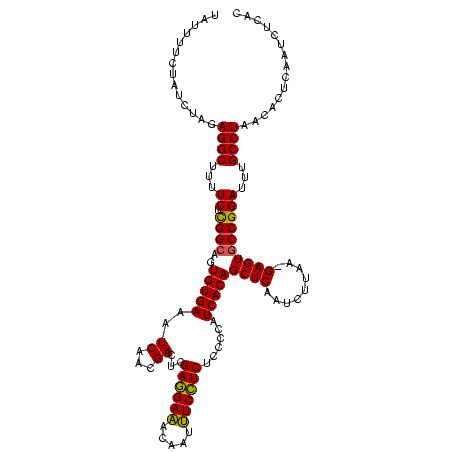
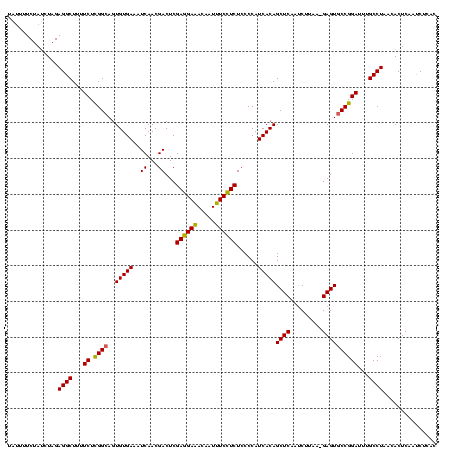
| Location | 2,495,471 – 2,495,590 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.64 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -24.79 |
| Energy contribution | -22.80 |
| Covariance contribution | -1.99 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.79 |
| Structure conservation index | 1.03 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 119 + 2685015/1360-1480 ACGACUCGAGGAAACAAUUUCCUCUCCCCAUCACAGCUCAAUCUUAA-GAGUGCCGGAUUUGCCUAACACUCAAUCUCACUGCUUGGACGUACAAUCCAAUCGUACGCUUCGCCUAUCCU .......((((((.....)))))).......................-(((((..((.....))...))))).........((..((.(((((.........)))))))..))....... ( -22.60) >sy_au.0 2231801 120 - 2809422/1360-1480 ACGACUCGAAGACACAAUGUCUUCUCCCCAUCACAGCUCAGCCUUAACGAGUACCGGAUUUGCCUAAUACUCAGCCUUACUGCUUAGACGUGCAAUCCAAUCGCACGCUUCGCCUAUCCU ...((((((((((.....))))))...........((...))......))))...((((..((......((.(((......))).)).(((((.........)))))....))..)))). ( -27.10) >sy_sa.0 2306606 119 + 2516575/1360-1480 ACGACUCGAGGAAACAAUUUCCUCUCCCCAUCACAGCUCAAUCUUAA-GAGUGCCGGAUUUGCCUAACACUCAAUCUCACUGCUUGGACGUACAAUCCAAUCGUACGCUUCGCCUAUCCU .......((((((.....)))))).......................-(((((..((.....))...))))).........((..((.(((((.........)))))))..))....... ( -22.60) >consensus ACGACUCGAGGAAACAAUUUCCUCUCCCCAUCACAGCUCAAUCUUAA_GAGUGCCGGAUUUGCCUAACACUCAAUCUCACUGCUUGGACGUACAAUCCAAUCGUACGCUUCGCCUAUCCU .......((((((.....))))))........................(((((..((.....))...))))).........((..((.(((((.........)))))))..))....... (-24.79 = -22.80 + -1.99)

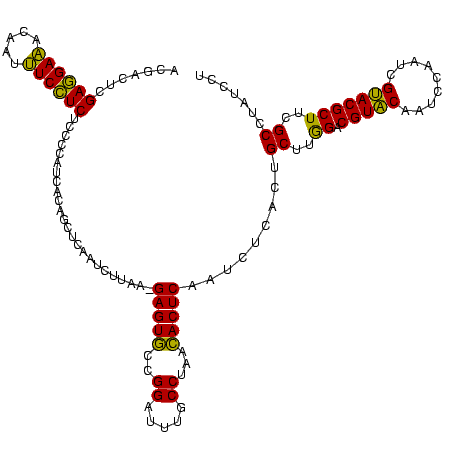
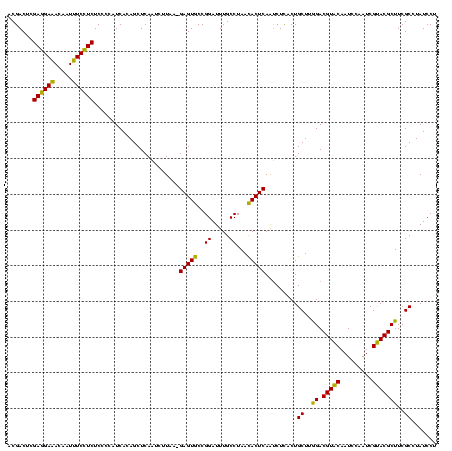
| Location | 2,495,471 – 2,495,590 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.64 |
| Mean single sequence MFE | -39.43 |
| Consensus MFE | -42.05 |
| Energy contribution | -39.17 |
| Covariance contribution | -2.88 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.46 |
| Structure conservation index | 1.07 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 119 + 2685015/1360-1480 AGGAUAGGCGAAGCGUACGAUUGGAUUGUACGUCCAAGCAGUGAGAUUGAGUGUUAGGCAAAUCCGGCACUC-UUAAGAUUGAGCUGUGAUGGGGAGAGGAAAUUGUUUCCUCGAGUCGU ......(((...(((((((((...)))))))))(((.(((((..((((((((((..((....))..))))))-....))))..)))))..)))...((((((.....))))))..))).. ( -39.20) >sy_au.0 2231801 120 - 2809422/1360-1480 AGGAUAGGCGAAGCGUGCGAUUGGAUUGCACGUCUAAGCAGUAAGGCUGAGUAUUAGGCAAAUCCGGUACUCGUUAAGGCUGAGCUGUGAUGGGGAGAAGACAUUGUGUCUUCGAGUCGU ......(((...(((((((((...)))))))))(((.(((((..((((((((((..((....))..)))))).....))))..)))))..)))...((((((.....))))))..))).. ( -39.90) >sy_sa.0 2306606 119 + 2516575/1360-1480 AGGAUAGGCGAAGCGUACGAUUGGAUUGUACGUCCAAGCAGUGAGAUUGAGUGUUAGGCAAAUCCGGCACUC-UUAAGAUUGAGCUGUGAUGGGGAGAGGAAAUUGUUUCCUCGAGUCGU ......(((...(((((((((...)))))))))(((.(((((..((((((((((..((....))..))))))-....))))..)))))..)))...((((((.....))))))..))).. ( -39.20) >consensus AGGAUAGGCGAAGCGUACGAUUGGAUUGUACGUCCAAGCAGUGAGAUUGAGUGUUAGGCAAAUCCGGCACUC_UUAAGAUUGAGCUGUGAUGGGGAGAGGAAAUUGUUUCCUCGAGUCGU ......(((...(((((((((...)))))))))(((.(((((..((((((((((..((....))..)))))).....))))..)))))..)))...((((((.....))))))..))).. (-42.05 = -39.17 + -2.88) # Strand winner: reverse (1.00)

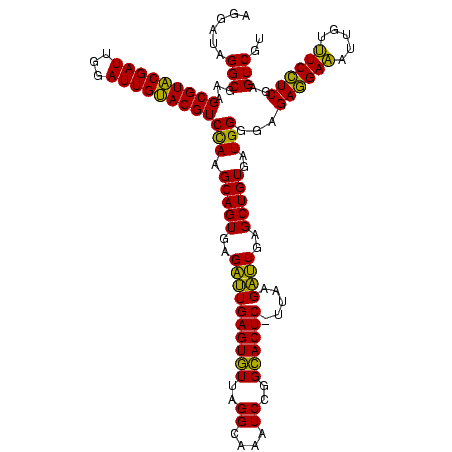
| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -44.73 |
| Consensus MFE | -41.97 |
| Energy contribution | -42.30 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 120 + 2685015/1480-1600 AAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGACAGGCGUAGGCGAUGGAUAACAGGUUGAUAUUCCUGUACCACCUAGCAUCGUUUUAAUCGAUGGGGGGACGCAGU ...((((((((.(((...(((((..((((((...(((.(.(((....))).).)))...)))).(((((........))))).))..)))))((((.......))))))).))))).))) ( -47.20) >sy_au.0 2231801 120 - 2809422/1480-1600 AAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGACAGGCGUAGGCGAUGGAUAACAGGUUGAUAUUCCUGUACCACCUAUAAUCGUUUUAAUCGAUGGGGGGACGCAGU ...((((((((((.(((.((..(((((..(((......))))))))..)).)))))..(((...(((((........))))).)))(((...((((.......)))).)))))))).))) ( -44.20) >sy_sa.0 2306606 120 + 2516575/1480-1600 AAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGAUAGGCGUAGGCGAUGGAUAACAGGUUGAUAUUCCUGUACCACCAUUAUUCGUUUUAAGCGAUGGGGGGACGCAGU ...((((((((.(((..(((.....((((((...(((.(.(((....))).).)))...)))).(((((........))))).))))).....((((.....)))).))).))))).))) ( -42.80) >consensus AAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGACAGGCGUAGGCGAUGGAUAACAGGUUGAUAUUCCUGUACCACCUAUAAUCGUUUUAAUCGAUGGGGGGACGCAGU ...((((((((.((((.((((((.(((((((...(((.(.(((....))).).)))...)))).(((((........)))))...........)))..)))))).).))).))))).))) (-41.97 = -42.30 + 0.33) # Strand winner: reverse (1.00)

| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -39.50 |
| Consensus MFE | -39.50 |
| Energy contribution | -39.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.09 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 120 + 2685015/1560-1680 CUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGC ....((((((((...((((((.((((((.(((......))))))))).....))))))..((((((((((((.......))))))....(.....).)))))).........)))))))) ( -39.50) >sy_au.0 2231801 120 - 2809422/1560-1680 CUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGC ....((((((((...((((((.((((((.(((......))))))))).....))))))..((((((((((((.......))))))....(.....).)))))).........)))))))) ( -39.50) >sy_sa.0 2306606 120 + 2516575/1560-1680 CUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGC ....((((((((...((((((.((((((.(((......))))))))).....))))))..((((((((((((.......))))))....(.....).)))))).........)))))))) ( -39.50) >consensus CUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGC ....((((((((...((((((.((((((.(((......))))))))).....))))))..((((((((((((.......))))))....(.....).)))))).........)))))))) (-39.50 = -39.50 + 0.00)

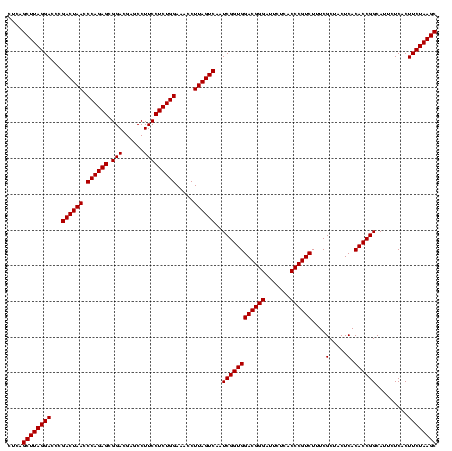
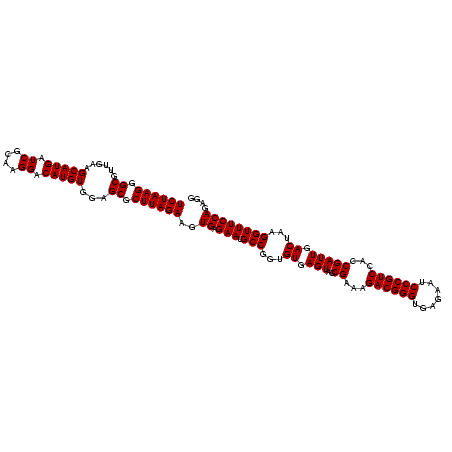
| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -37.97 |
| Consensus MFE | -37.97 |
| Energy contribution | -37.97 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.38 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 120 + 2685015/1600-1720 UCUAAGGGCGUUGAAGCAUGAUCGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGAUUGACUAAGGUUUCCAGAGG ((((((.((......(((((.((....)).)))))...)).))))))..((.(((.(((...((.(((..((...((((((.......))))))...))))).))...)))))))).... ( -38.30) >sy_au.0 2231801 120 - 2809422/1600-1720 UCUAAGGGCGUUGAAGCAUGAUCGUAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGAUUGACUAAGGUUUCCAGAGG ((((((.((......(((((.((....)).)))))...)).))))))..((.(((.(((...((.(((..((...((((((.......))))))...))))).))...)))))))).... ( -37.30) >sy_sa.0 2306606 120 + 2516575/1600-1720 UCUAAGGGCGUUGAAGCAUGAUCGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGAUUGACUAAGGUUUCCAGAGG ((((((.((......(((((.((....)).)))))...)).))))))..((.(((.(((...((.(((..((...((((((.......))))))...))))).))...)))))))).... ( -38.30) >consensus UCUAAGGGCGUUGAAGCAUGAUCGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGAUUGACUAAGGUUUCCAGAGG ((((((.((......(((((.((....)).)))))...)).))))))..((.(((.(((...((.(((..((...((((((.......))))))...))))).))...)))))))).... (-37.97 = -37.97 + -0.00) # Strand winner: reverse (1.00)

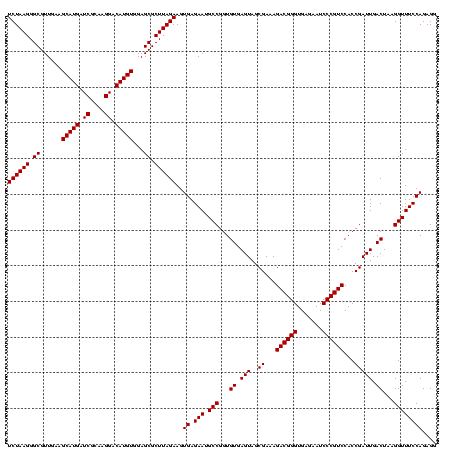
| Location | 2,495,471 – 2,495,590 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.77 |
| Mean single sequence MFE | -20.41 |
| Consensus MFE | -21.44 |
| Energy contribution | -21.44 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 1.05 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799716 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
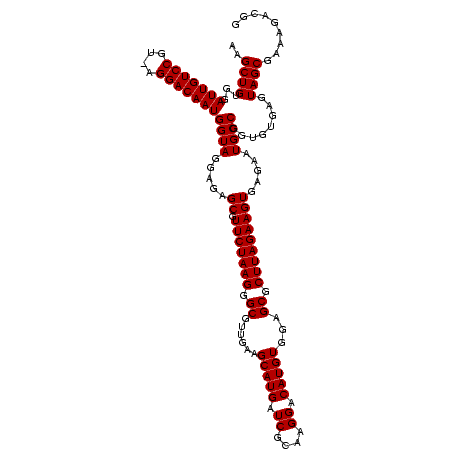
>sy_ha.0 2495471 119 + 2685015/1640-1760 CCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUGCGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCCU-ACGGACAAUCCACAGCUU .........(((.........((.........(((((((.((....((((((.....)).)))).......)).)))))))..........))(((((((.-..)))))))....))).. ( -19.91) >sy_au.0 2231801 119 - 2809422/1640-1760 CCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUACGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCCA-AAGGACAAUCCACAGCUU .........(((.........((.........(((((((.((....((((((.....)).)))).......)).)))))))..........))(((((((.-..)))))))....))).. ( -19.91) >sy_sa.0 2306606 120 + 2516575/1640-1760 CCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUGCGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCCUUACGGACAAUCCACAGCUU .........(((.........((.........(((((((.((....((((((.....)).)))).......)).)))))))..........))(((((((....)))))))....))).. ( -21.41) >consensus CCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUGCGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCCU_ACGGACAAUCCACAGCUU .........(((.........((.........(((((((.((....((((((.....)).)))).......)).)))))))..........))(((((((....)))))))....))).. (-21.44 = -21.44 + 0.00)

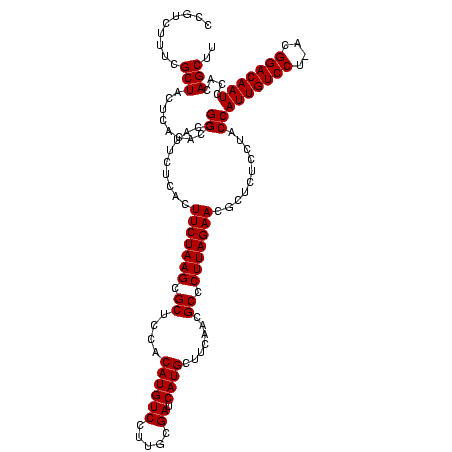
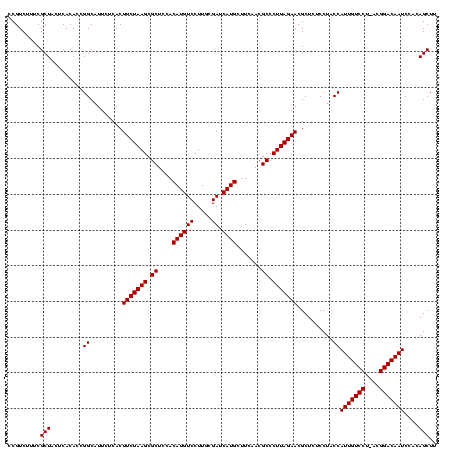
| Location | 2,495,471 – 2,495,590 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.77 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -41.37 |
| Energy contribution | -41.37 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.60 |
| Structure conservation index | 1.04 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 119 + 2685015/1640-1760 AAGCUGUGGAUUGUCCGU-AGGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGG ..((((...(((((((..-.)))))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))........))))......... ( -38.80) >sy_au.0 2231801 119 - 2809422/1640-1760 AAGCUGUGGAUUGUCCUU-UGGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGUAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGG ..((((...(((((((..-.)))))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))........))))......... ( -37.80) >sy_sa.0 2306606 120 + 2516575/1640-1760 AAGCUGUGGAUUGUCCGUAAGGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGG ..((((...(((((((....)))))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))........))))......... ( -43.10) >consensus AAGCUGUGGAUUGUCCGU_AGGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGG ..((((...(((((((....)))))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))........))))......... (-41.37 = -41.37 + 0.00) # Strand winner: reverse (1.00)

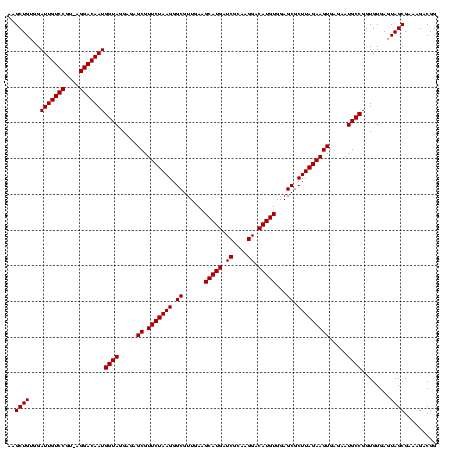
| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -35.04 |
| Consensus MFE | -35.34 |
| Energy contribution | -35.23 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.39 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 120 + 2685015/1880-2000 CCGAAUGCCAAUUAAUUUAACUUGGGAGUCAGAACAUGGGUGAUAAGGUCCGUGUUCGAAAGGGAAACAGCCCAGACCACCAGCUAAGGUCCCAAAAUAUAUGUUAAGUGGAAAAGGAUG ((.....(((.(((((.((..(((((((((.(((((((((........)))))))))....(((......))).)))..((......))))))))..))...))))).)))....))... ( -35.50) >sy_au.0 2231801 120 - 2809422/1880-2000 CCGAAUGCCAAUUAAUUUAACUUGGGAGUCAGAACAUGGGUGAUAAGGUCCGUGUUCGAAAGGGAAACAGCCCAGACCACCAGCUAAGGUCCCAAAAUAUAUGUUAAGUGGAAAAGGAUG ((.....(((.(((((.((..(((((((((.(((((((((........)))))))))....(((......))).)))..((......))))))))..))...))))).)))....))... ( -35.50) >sy_sa.0 2306606 120 + 2516575/1880-2000 CCGAAUGCCAAUCAAUUUAACUUGGGAGUCAGAACGCGGGUGAUAAGGUCCGUGUUCGAAAGGGAAACAGCCCAGACCACCAGCUAAGGUCCCAAAAUAUAUGUUAAGUGGAAAAGGAUG .......((..((.((((((((((((((((.(((((((((........)))))))))....(((......))).)))..((......)))))))).......))))))).))...))... ( -34.11) >consensus CCGAAUGCCAAUUAAUUUAACUUGGGAGUCAGAACAUGGGUGAUAAGGUCCGUGUUCGAAAGGGAAACAGCCCAGACCACCAGCUAAGGUCCCAAAAUAUAUGUUAAGUGGAAAAGGAUG ((.....(((.(((((.((..(((((((((.(((((((((........)))))))))....(((......))).)))..((......))))))))..))...))))).)))....))... (-35.34 = -35.23 + -0.11) # Strand winner: reverse (0.96)

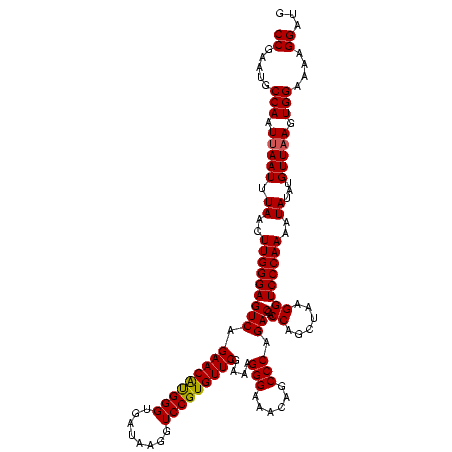

| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -39.57 |
| Consensus MFE | -37.91 |
| Energy contribution | -37.80 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.96 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 120 + 2685015/2000-2120 ACUUGGAGAUAGCUGGUUCUCUCCGAAAUAGCUUUAGGGCUAGCCUCAAGUGAUGAUUAUUGGAGGUAGAGCACUGUUUGGACGAGGGGCCCCUCUCGGGUUACCGAAUUCAGACAAACU ..(((((((.((....)).)))))))..(((((....)))))(((((.(((((...))))).))))).......((((((((.((((....))))((((....)))).)))))))).... ( -39.50) >sy_au.0 2231801 120 - 2809422/2000-2120 ACCUGGAGAUAGCUGGUUCUCUCCGAAAUAGCUUUAGGGCUAGCCUCAAGUGAUGAUUAUUGGAGGUAGAGCACUGUUUGGACGAGGGGCCCCUCUCGGGUUACCGAAUUCAGACAAACU .(((((((.((..(((......)))...)).)))))))(((.(((((.(((((...))))).)))))..)))..((((((((.((((....))))((((....)))).)))))))).... ( -41.00) >sy_sa.0 2306606 120 + 2516575/2000-2120 ACCUGGAGAUAGCUGGUUCUCUCCGAAAUAGCUUUAGGGCUAGCCUCAAGUGAUGAUUAUUGGAGGUAGAGCACUGUUUGGACGAGGGGCCCUUAUCGGGUUACCGAAUUCAGACAAACU .(((((((.((..(((......)))...)).)))))))(((.(((((.(((((...))))).)))))..)))..((((((((...(((((((.....))))).))...)))))))).... ( -38.20) >consensus ACCUGGAGAUAGCUGGUUCUCUCCGAAAUAGCUUUAGGGCUAGCCUCAAGUGAUGAUUAUUGGAGGUAGAGCACUGUUUGGACGAGGGGCCCCUCUCGGGUUACCGAAUUCAGACAAACU .(((((((.((..(((......)))...)).)))))))(((.(((((.(((((...))))).)))))..)))..((((((((.((((....))))((((....)))).)))))))).... (-37.91 = -37.80 + -0.11) # Strand winner: reverse (1.00)

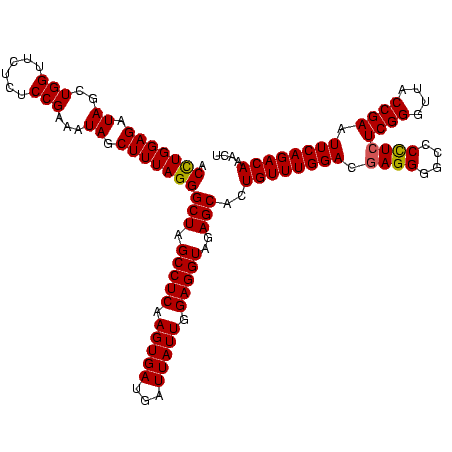

| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -41.57 |
| Consensus MFE | -42.86 |
| Energy contribution | -41.53 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.44 |
| Structure conservation index | 1.03 |
| SVM decision value | 2.80 |
| SVM RNA-class probability | 0.997133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 120 + 2685015/2120-2240 GAAACCAGGUGAUCUACCCAUGGUCAGGUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAACUGUGGGUAGCGGAGAAAUUCCAAUCGA .......(((...(((((((((((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))))))))))).((((...)))).))).. ( -41.40) >sy_au.0 2231801 120 - 2809422/2120-2240 GAAACCAGGUGAUCUACCCUUGGUCAGGUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAACUGAGGGUAGCGGAGAAAUUCCAAUCGA .......(((...(((((((..((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))..))))))).((((...)))).))).. ( -41.70) >sy_sa.0 2306606 120 + 2516575/2120-2240 GAAACCAGGUGAUCUACCCAUGACCAGGUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAGUUGUGGGUAGCGGAGAAAUUCCAAUCGA .......(((...(((((((..((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))..))))))).((((...)))).))).. ( -41.60) >consensus GAAACCAGGUGAUCUACCCAUGGUCAGGUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAACUGUGGGUAGCGGAGAAAUUCCAAUCGA .......(((...(((((((..((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))..))))))).((((...)))).))).. (-42.86 = -41.53 + -1.33) # Strand winner: reverse (0.99)

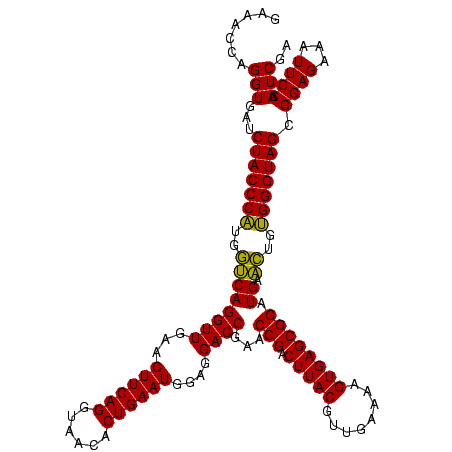
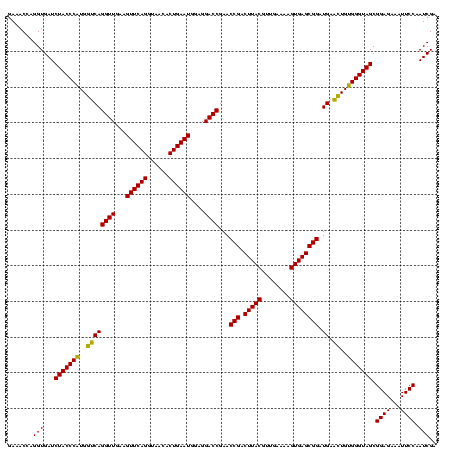
| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -22.57 |
| Consensus MFE | -23.38 |
| Energy contribution | -23.17 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.22 |
| Structure conservation index | 1.04 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 120 + 2685015/2520-2640 AUUUCACGUGCUCCGUCGUACUCAGGAUCCACUCAAGAGAGAAUAUAUUUUCAACUACAGGAUUAUUACCUUCUUUGAUUCAACUUUCCAGAUGAUUCGUCUAAUAUAUUCCUUUGUAAC .......(((.(((..........)))..)))...((((.(((((((((..........((((((..........))))))........(((((...))))))))))))))))))..... ( -18.60) >sy_au.0 2231801 120 - 2809422/2520-2640 AUUUCACGUGCUCCGUCGUACUCAGGAUCCACUCAAGAGAGACAACAUUUUCGACUACAGGAUUAUUACCUUCUUUGAUUCAUCUUUCCAGAUGAUUCGUCUAAUGUCGUCCUUUGUAAC .......(((.(((..........)))..)))...((((.(((.(((((...(((...(((.......))).....((.((((((....)))))).))))).))))).)))))))..... ( -25.10) >sy_sa.0 2306606 120 + 2516575/2520-2640 AUUUCUCGUGUUCCGUCGUACUCAGGAUCCACUCAAGAGUGAACAAACUUUCGACUACAGGAUUAUUACCUUCUUUGAUUCAUCUUUCCAGAUGAUUCGUCUAGUCUGUUCUUUUGUAAC .......(((.(((..........)))..))).(((((..(((((.(((...(((...(((.......))).....((.((((((....)))))).))))).))).)))))))))).... ( -24.00) >consensus AUUUCACGUGCUCCGUCGUACUCAGGAUCCACUCAAGAGAGAAAAAAUUUUCGACUACAGGAUUAUUACCUUCUUUGAUUCAUCUUUCCAGAUGAUUCGUCUAAUAUAUUCCUUUGUAAC .......(((.(((..........)))..)))...((((.(((((((((...(((...(((.......))).....((.((((((....)))))).))))).)))))))))))))..... (-23.38 = -23.17 + -0.21)

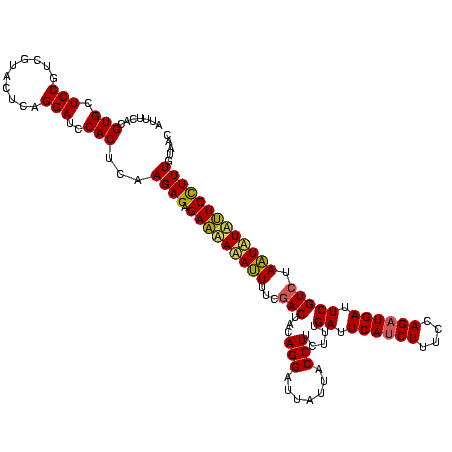
| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -30.63 |
| Consensus MFE | -31.61 |
| Energy contribution | -29.83 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.30 |
| Structure conservation index | 1.03 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994526 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 120 + 2685015/2520-2640 GUUACAAAGGAAUAUAUUAGACGAAUCAUCUGGAAAGUUGAAUCAAAGAAGGUAAUAAUCCUGUAGUUGAAAAUAUAUUCUCUCUUGAGUGGAUCCUGAGUACGACGGAGCACGUGAAAU .((((...((((((((((.(((((.(((.((....)).))).)).....(((.......)))...)))...)))))))))).......(((..(((..........))).)))))))... ( -24.60) >sy_au.0 2231801 120 - 2809422/2520-2640 GUUACAAAGGACGACAUUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCGAAAAUGUUGUCUCUCUUGAGUGGAUCCUGAGUACGACGGAGCACGUGAAAU .((((...((((((((((.(((((.((((((....)))))).)).....(((.......)))...)))...)))))))))).......(((..(((..........))).)))))))... ( -34.40) >sy_sa.0 2306606 120 + 2516575/2520-2640 GUUACAAAAGAACAGACUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCGAAAGUUUGUUCACUCUUGAGUGGAUCCUGAGUACGACGGAACACGAGAAAU .........(((((((((.(((((.((((((....)))))).)).....(((.......)))...)))...)))))))))..((((..(((..(((..........))).)))))))... ( -32.90) >consensus GUUACAAAGGAACACAUUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCGAAAAUAUAUUCUCUCUUGAGUGGAUCCUGAGUACGACGGAGCACGUGAAAU .((((....(((((((((.(((((.((((((....)))))).)).....(((.......)))...)))...)))))))))........(((..(((..........))).)))))))... (-31.61 = -29.83 + -1.77) # Strand winner: reverse (1.00)

| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -21.87 |
| Consensus MFE | -23.20 |
| Energy contribution | -23.10 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.42 |
| Structure conservation index | 1.06 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 120 + 2685015/2560-2680 GAAUAUAUUUUCAACUACAGGAUUAUUACCUUCUUUGAUUCAACUUUCCAGAUGAUUCGUCUAAUAUAUUCCUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAAUAAGCAAGCUUAU (((((((((..........((((((..........))))))........(((((...))))))))))))))..(((((((((......))))....))))).....(((((....))))) ( -17.30) >sy_au.0 2231801 120 - 2809422/2560-2680 GACAACAUUUUCGACUACAGGAUUAUUACCUUCUUUGAUUCAUCUUUCCAGAUGAUUCGUCUAAUGUCGUCCUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAACAAGCAAGCUUGU (((.(((((...(((...(((.......))).....((.((((((....)))))).))))).))))).)))..(((((((((......))))....))))).....(((((....))))) ( -25.70) >sy_sa.0 2306606 120 + 2516575/2560-2680 GAACAAACUUUCGACUACAGGAUUAUUACCUUCUUUGAUUCAUCUUUCCAGAUGAUUCGUCUAGUCUGUUCUUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAACAAGCAAGCUCGU (((((.(((...(((...(((.......))).....((.((((((....)))))).))))).))).)))))..(((((((((......))))....)))))................... ( -22.60) >consensus GAAAAAAUUUUCGACUACAGGAUUAUUACCUUCUUUGAUUCAUCUUUCCAGAUGAUUCGUCUAAUAUAUUCCUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAACAAGCAAGCUUGU (((((((((...(((...(((.......))).....((.((((((....)))))).))))).)))))))))..(((((((((......))))....))))).....(((((....))))) (-23.20 = -23.10 + -0.10)

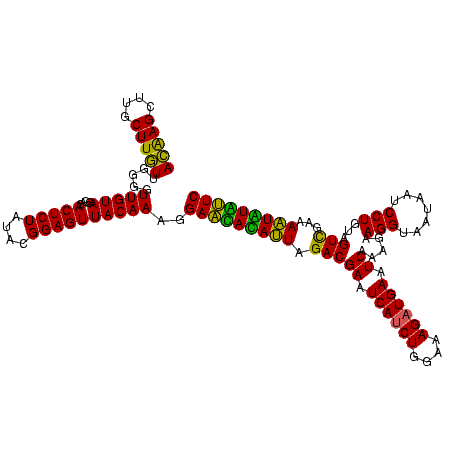
| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -30.53 |
| Consensus MFE | -33.64 |
| Energy contribution | -30.87 |
| Covariance contribution | -2.77 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.06 |
| Structure conservation index | 1.10 |
| SVM decision value | 2.96 |
| SVM RNA-class probability | 0.997910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 120 + 2685015/2560-2680 AUAAGCUUGCUUAUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGAAUAUAUUAGACGAAUCAUCUGGAAAGUUGAAUCAAAGAAGGUAAUAAUCCUGUAGUUGAAAAUAUAUUC (((((....))))).....(((((....(((((....))))))))))..(((((((((.(((((.(((.((....)).))).)).....(((.......)))...)))...))))))))) ( -22.70) >sy_au.0 2231801 120 - 2809422/2560-2680 ACAAGCUUGCUUGUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGACGACAUUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCGAAAAUGUUGUC (((((....))))).....(((((....(((((....))))))))))..(((((((((.(((((.((((((....)))))).)).....(((.......)))...)))...))))))))) ( -34.40) >sy_sa.0 2306606 120 + 2516575/2560-2680 ACGAGCUUGCUUGUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAAGAACAGACUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCGAAAGUUUGUUC (((((....))))).....(((((....(((((....))))))))))..(((((((((.(((((.((((((....)))))).)).....(((.......)))...)))...))))))))) ( -34.50) >consensus ACAAGCUUGCUUGUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGAACACAUUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCGAAAAUAUAUUC (((((....))))).....(((((....(((((....))))))))))..(((((((((.(((((.((((((....)))))).)).....(((.......)))...)))...))))))))) (-33.64 = -30.87 + -2.77) # Strand winner: reverse (1.00)


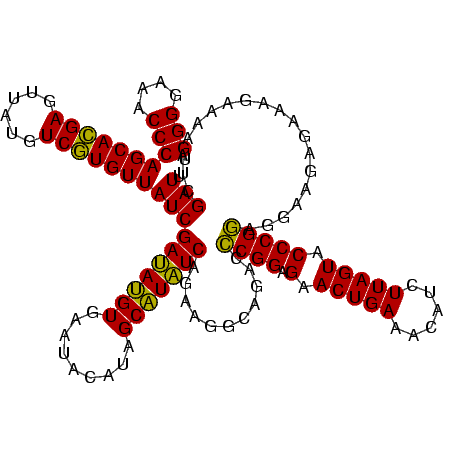
| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -35.14 |
| Energy contribution | -33.37 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.83 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 120 + 2685015/2720-2840 UGGGGGAACCCAGCACGAGUUAUGUCGUGUUAUCGACAAGUGAAUUCAUAGCUUGUCAGAAGGCAGACCCGGAGAACUGAAACAUCUUAGUACCCGGAGGAAGAGAAAGAAAAAUCGAUU ((((....))))...(((.....(((.(((..(((((((((.........))))))).))..))))))((((.(.(((((......))))).))))).................)))... ( -35.10) >sy_au.0 2231801 120 - 2809422/2720-2840 UGGGGAAACCCAGCAUGAGUUAUGUCAUGUUAUCGAUAUGUGAAUACAUAGCAUAUCAGAAGGCACACCCGGAGAACUGAAACAUCUUAGUACCCGGAGGAAGAGAAAGAAAAUUCGAUU ((((....))))...((((((...((.(((..(((((((((.........))))))).))..)))(..((((.(.(((((......))))).)))))..)........)).))))))... ( -34.30) >sy_sa.0 2306606 120 + 2516575/2720-2840 UGGGGAAACCCAGCACGAGUUAUGUCGUGUUAUCGAUAUGUGAAUACAUAGCAUAUCUGAAGGCAGACGCGGAGAACUGAAACAUCUUAGUACCCGCAGGAAGAGAAAGAAAAAUCGAUU ((((....))))...(((.....(((.(((..(((((((((.........))))))).))..))))))((((.(.(((((......))))).))))).................)))... ( -36.50) >consensus UGGGGAAACCCAGCACGAGUUAUGUCGUGUUAUCGAUAUGUGAAUACAUAGCAUAUCAGAAGGCAGACCCGGAGAACUGAAACAUCUUAGUACCCGGAGGAAGAGAAAGAAAAAUCGAUU .(((....)))(((((((......)))))))((((((((((.........)))))))...........((((.(.(((((......))))).)))))...................))). (-35.14 = -33.37 + -1.77) # Strand winner: reverse (1.00)


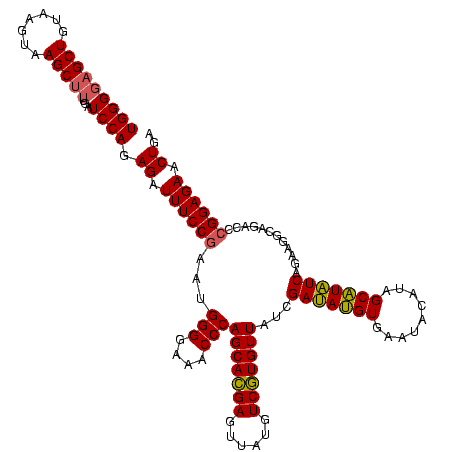
| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -35.10 |
| Energy contribution | -34.77 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 120 + 2685015/2760-2880 UGGGUAGCUGUAAGUAAGCGUUGAUCCAGAGAUUUCCGAAUGGGGGAACCCAGCACGAGUUAUGUCGUGUUAUCGACAAGUGAAUUCAUAGCUUGUCAGAAGGCAGACCCGGAGAACUGA (((((.(((.......)))....))))).((.((((((..((((....))))...........(((.(((..(((((((((.........))))))).))..)))))).)))))).)).. ( -38.70) >sy_au.0 2231801 120 - 2809422/2760-2880 UGGGGAGCUGUAAGUAAGCUUUGAUCCAGAGAUUUCCGAAUGGGGAAACCCAGCAUGAGUUAUGUCAUGUUAUCGAUAUGUGAAUACAUAGCAUAUCAGAAGGCACACCCGGAGAACUGA (((((((((.......)))))...)))).((.((((((...(((....)))(((((((......)))))))...(((((((.........)))))))............)))))).)).. ( -36.50) >sy_sa.0 2306606 120 + 2516575/2760-2880 UGGGGAGCUGUAAGUAAGCUUUGAUCCAGAGAUUUCCUAAUGGGGAAACCCAGCACGAGUUAUGUCGUGUUAUCGAUAUGUGAAUACAUAGCAUAUCUGAAGGCAGACGCGGAGAACUGA (((((((((.......)))))...)))).((.(((((...((((....))))...........(((.(((..(((((((((.........))))))).))..))))))..))))).)).. ( -36.70) >consensus UGGGGAGCUGUAAGUAAGCUUUGAUCCAGAGAUUUCCGAAUGGGGAAACCCAGCACGAGUUAUGUCGUGUUAUCGAUAUGUGAAUACAUAGCAUAUCAGAAGGCAGACCCGGAGAACUGA (((((((((.......)))))...)))).((.((((((...(((....)))(((((((......)))))))...(((((((.........)))))))............)))))).)).. (-35.10 = -34.77 + -0.33) # Strand winner: reverse (1.00)

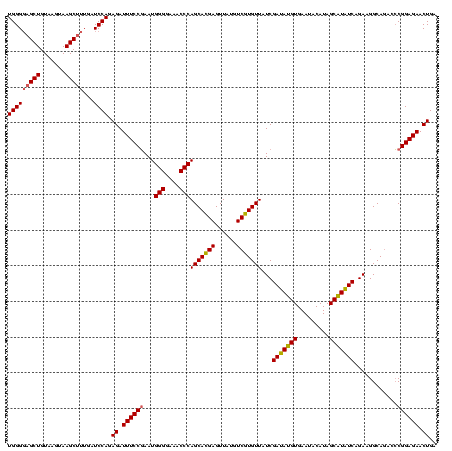
| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.93 |
| Mean single sequence MFE | -27.37 |
| Consensus MFE | -18.14 |
| Energy contribution | -18.14 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.90 |
| SVM RNA-class probability | 0.997633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 120 + 2685015/2920-3040 UGCCAAGGCAUCCACCGUGCGCCCUUAAUAACUUAAUCUAUGUUUCAACCAUAUUAGAACGUAGUUCUAAUUUUAAGAAACCUGUUAUUAAUCUUGUGAGUGUUCUUUCGAACACUAGCG .((((((((((.....))))....((((((((.........(((((......((((((((...)))))))).....)))))..)))))))).))))..(((((((....))))))).)). ( -28.10) >sy_au.0 2231801 100 - 2809422/2920-3040 UGCCAAGGCAUCCACCGUGCGCCCUUAAUAACUUAAUCUAUGUUUCCAUCCUAC--------------------AGGAAACGCGUUAUUAAUCUUGUGAGUGUUCUUUCGAACACUAGCG .((((((((((.....))))....((((((((........(((((((.......--------------------.))))))).)))))))).))))..(((((((....))))))).)). ( -25.40) >sy_sa.0 2306606 118 + 2516575/2920-3040 UGCCAAGGCAUCCACCGUGCGCCCUUAAUAACUUAAUCUUUUUUGACUUUCAACACGAACA-AGUCGGUUGAAAACCCAAAAUGUUAUUAAUCU-GUGAGUGUUCUUUCGAACACUAGCG (((....))).....(((.(((..((((((((........(((((..(((((((.(((...-..))))))))))...))))).))))))))...-)))(((((((....))))))).))) ( -28.60) >consensus UGCCAAGGCAUCCACCGUGCGCCCUUAAUAACUUAAUCUAUGUUUCCAUCCUAC__GAAC__AGU_________AAGAAACAUGUUAUUAAUCUUGUGAGUGUUCUUUCGAACACUAGCG .((((((((((.....))))....((((((((........((((((..............................)))))).)))))))).))))..(((((((....))))))).)). (-18.14 = -18.14 + 0.00)

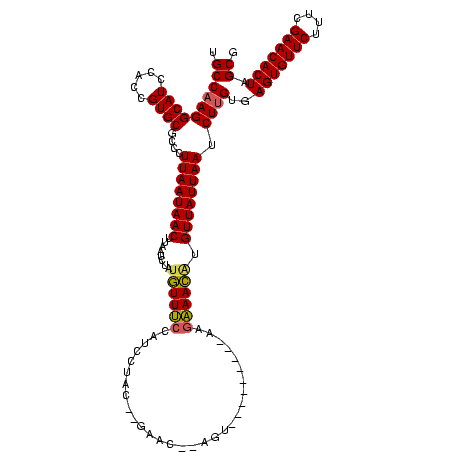
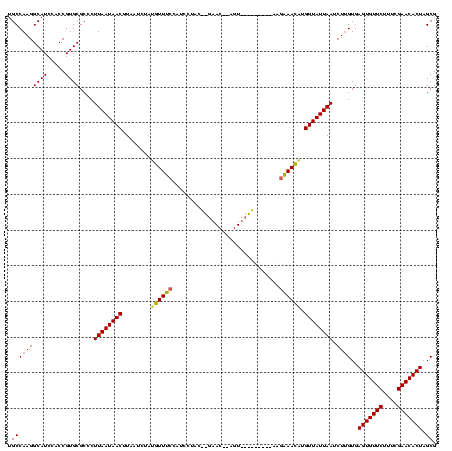
| Location | 2,495,471 – 2,495,591 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.93 |
| Mean single sequence MFE | -33.93 |
| Consensus MFE | -26.27 |
| Energy contribution | -25.23 |
| Covariance contribution | -1.05 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.77 |
| SVM decision value | 4.71 |
| SVM RNA-class probability | 0.999942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 120 + 2685015/2920-3040 CGCUAGUGUUCGAAAGAACACUCACAAGAUUAAUAACAGGUUUCUUAAAAUUAGAACUACGUUCUAAUAUGGUUGAAACAUAGAUUAAGUUAUUAAGGGCGCACGGUGGAUGCCUUGGCA .((((((((((....))))))).......((((((((..(((((.....((((((((...))))))))......))))).........)))))))).)))((..(((....)))...)). ( -33.10) >sy_au.0 2231801 100 - 2809422/2920-3040 CGCUAGUGUUCGAAAGAACACUCACAAGAUUAAUAACGCGUUUCCU--------------------GUAGGAUGGAAACAUAGAUUAAGUUAUUAAGGGCGCACGGUGGAUGCCUUGGCA .((((((((((....))))))).......((((((((.....(((.--------------------...)))((....))........)))))))).)))((..(((....)))...)). ( -30.80) >sy_sa.0 2306606 118 + 2516575/2920-3040 CGCUAGUGUUCGAAAGAACACUCAC-AGAUUAAUAACAUUUUGGGUUUUCAACCGACU-UGUUCGUGUUGAAAGUCAAAAAAGAUUAAGUUAUUAAGGGCGCACGGUGGAUGCCUUGGCA .((((((((((....)))))))...-...(((((((((((((.(..((((((((((..-...))).)))))))..)....)))))...)))))))).)))((..(((....)))...)). ( -37.90) >consensus CGCUAGUGUUCGAAAGAACACUCACAAGAUUAAUAACACGUUUCCU_________ACU__GUUC__GUAGGAUGGAAACAUAGAUUAAGUUAUUAAGGGCGCACGGUGGAUGCCUUGGCA .((((((((((....))))))).......((((((((..(((((.......((((((...))))))........))))).........)))))))).)))((..(((....)))...)). (-26.27 = -25.23 + -1.05) # Strand winner: reverse (1.00)

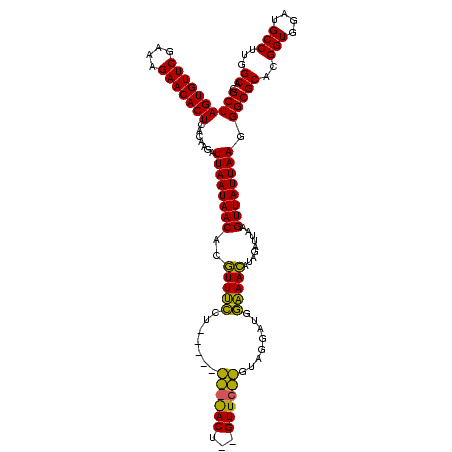
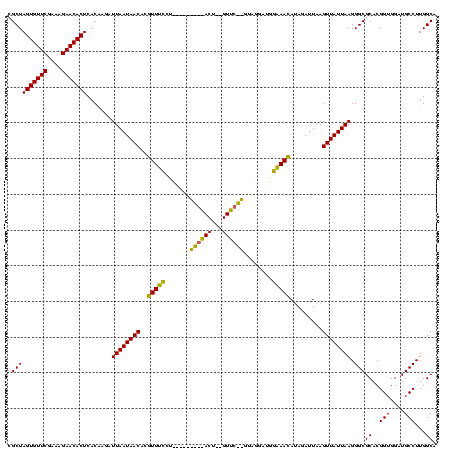
| Location | 2,495,471 – 2,495,588 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.07 |
| Mean single sequence MFE | -23.77 |
| Consensus MFE | -12.60 |
| Energy contribution | -12.04 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.53 |
| SVM decision value | 3.45 |
| SVM RNA-class probability | 0.999225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 117 + 2685015/2960-3080 UGUUUCAACCAUAUUAGAACGUAGUUCUAAUUUUAAGAAACCUGUUAUUAAUCUUGUGAGUGUUCUUUCGAACACUAGCGAUUAUAUUUUAU-GAAUUCAA-GCUUUUUUAAAACUCUU- .(((((......((((((((...)))))))).....))))).......(((((..((.(((((((....))))))).)))))))..(((((.-(((.....-...))).))))).....- ( -21.30) >sy_au.0 2231801 98 - 2809422/2960-3080 UGUUUCCAUCCUAC--------------------AGGAAACGCGUUAUUAAUCUUGUGAGUGUUCUUUCGAACACUAGCGAUUAUUUCUUAU-GAAUUCAA-GCUUAUUUAAAACUCUUU .((((((.......--------------------.))))))((.....(((((..((.(((((((....))))))).))))))).(((....-))).....-))................ ( -21.20) >sy_sa.0 2306606 118 + 2516575/2960-3080 UUUUGACUUUCAACACGAACA-AGUCGGUUGAAAACCCAAAAUGUUAUUAAUCU-GUGAGUGUUCUUUCGAACACUAGCGAUUAUUUCUUUUUGAAUUCAAAGCUUGUUAAAAAACUCUA ((((((((((((((.(((...-..))))))))))...(((((.(....(((((.-((.(((((((....))))))).)))))))...).)))))............)))))))....... ( -28.80) >consensus UGUUUCCAUCCUAC__GAAC__AGU_________AAGAAACAUGUUAUUAAUCUUGUGAGUGUUCUUUCGAACACUAGCGAUUAUUUCUUAU_GAAUUCAA_GCUUAUUUAAAACUCUU_ .(((((..............................))))).......(((((..((.(((((((....))))))).))))))).................................... (-12.60 = -12.04 + -0.55)

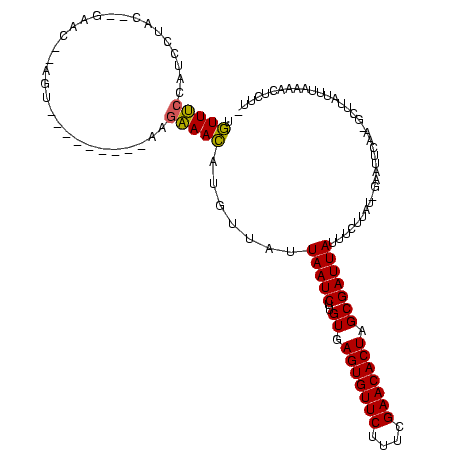
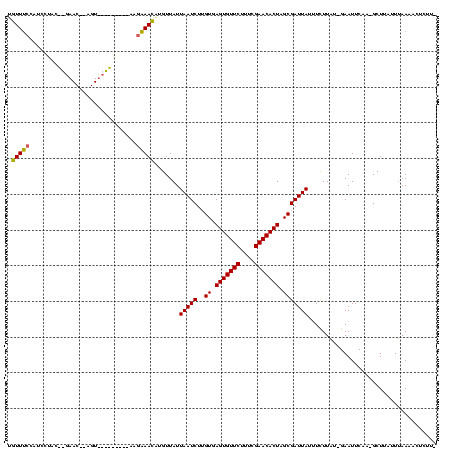
| Location | 2,495,471 – 2,495,588 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.07 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -18.57 |
| Energy contribution | -17.53 |
| Covariance contribution | -1.05 |
| Combinations/Pair | 1.26 |
| Mean z-score | -4.19 |
| Structure conservation index | 0.64 |
| SVM decision value | 7.15 |
| SVM RNA-class probability | 1.000000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2495471 117 + 2685015/2960-3080 -AAGAGUUUUAAAAAAGC-UUGAAUUC-AUAAAAUAUAAUCGCUAGUGUUCGAAAGAACACUCACAAGAUUAAUAACAGGUUUCUUAAAAUUAGAACUACGUUCUAAUAUGGUUGAAACA -....(((((((..((((-(((.(((.-....))).(((((...(((((((....))))))).....)))))....)))))))......((((((((...))))))))....))))))). ( -28.40) >sy_au.0 2231801 98 - 2809422/2960-3080 AAAGAGUUUUAAAUAAGC-UUGAAUUC-AUAAGAAAUAAUCGCUAGUGUUCGAAAGAACACUCACAAGAUUAAUAACGCGUUUCCU--------------------GUAGGAUGGAAACA ...((((((.....))))-))......-........(((((...(((((((....))))))).....))))).......((((((.--------------------.......)))))). ( -26.10) >sy_sa.0 2306606 118 + 2516575/2960-3080 UAGAGUUUUUUAACAAGCUUUGAAUUCAAAAAGAAAUAAUCGCUAGUGUUCGAAAGAACACUCAC-AGAUUAAUAACAUUUUGGGUUUUCAACCGACU-UGUUCGUGUUGAAAGUCAAAA ((((((((......))))))))....(((((.(...(((((...(((((((....)))))))...-.)))))....).)))))(..((((((((((..-...))).)))))))..).... ( -32.70) >consensus _AAGAGUUUUAAAAAAGC_UUGAAUUC_AUAAGAAAUAAUCGCUAGUGUUCGAAAGAACACUCACAAGAUUAAUAACACGUUUCCU_________ACU__GUUC__GUAGGAUGGAAACA ....................................(((((...(((((((....))))))).....))))).......(((((.......((((((...))))))........))))). (-18.57 = -17.53 + -1.05) # Strand winner: reverse (1.00)

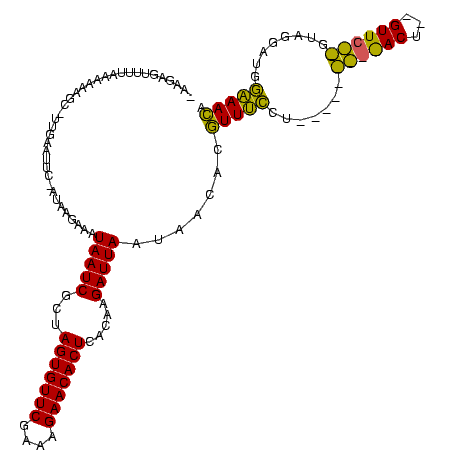
| Location | 2,495,471 – 2,495,588 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.05 |
| Mean single sequence MFE | -28.10 |
| Consensus MFE | -15.11 |
| Energy contribution | -15.00 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -4.10 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
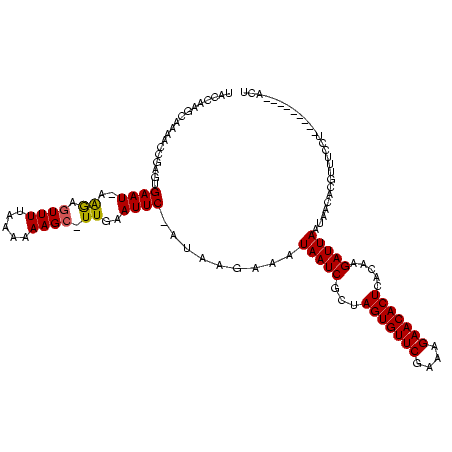
>sy_ha.0 2495471 117 + 2685015/2982-3102 UACCAAGCAAAACCGAGUGAAU-AAGAGUUUUAAAAAAGC-UUGAAUUC-AUAAAAUAUAAUCGCUAGUGUUCGAAAGAACACUCACAAGAUUAAUAACAGGUUUCUUAAAAUUAGAACU ....(((..(((((..((((((-..((((((.....))))-))..))))-))......(((((...(((((((....))))))).....)))))......))))))))............ ( -28.50) >sy_au.0 2231801 106 - 2809422/2982-3102 UACCAAGCAAAACCGAGUGAAUAAAGAGUUUUAAAUAAGC-UUGAAUUC-AUAAGAAAUAAUCGCUAGUGUUCGAAAGAACACUCACAAGAUUAAUAACGCGUUUCCU------------ .........((((((.((((((...((((((.....))))-))..))))-))......(((((...(((((((....))))))).....)))))....)).))))...------------ ( -26.10) >sy_sa.0 2306606 119 + 2516575/2982-3102 UACCAAGCAAAACCGAGUGAAUUAGAGUUUUUUAACAAGCUUUGAAUUCAAAAAGAAAUAAUCGCUAGUGUUCGAAAGAACACUCAC-AGAUUAAUAACAUUUUGGGUUUUCAACCGACU ......(.((((((.(((((((((((((((......)))))))).)))))........(((((...(((((((....)))))))...-.)))))........)).)))))))........ ( -29.70) >consensus UACCAAGCAAAACCGAGUGAAU_AAGAGUUUUAAAAAAGC_UUGAAUUC_AUAAGAAAUAAUCGCUAGUGUUCGAAAGAACACUCACAAGAUUAAUAACACGUUUCCU_________ACU ..................((((..((.((((.....)))).))..)))).........(((((...(((((((....))))))).....))))).......................... (-15.11 = -15.00 + -0.11) # Strand winner: reverse (0.69)


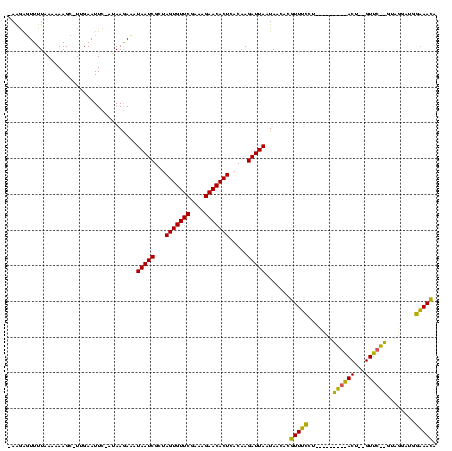
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:57:06 2006