| Sequence ID | sy_ha.0 |
|---|---|
| Location | 1,799,678 – 1,799,759 |
| Length | 81 |
| Max. P | 0.998353 |

| Location | 1,799,678 – 1,799,759 |
|---|---|
| Length | 81 |
| Sequences | 5 |
| Columns | 81 |
| Reading direction | forward |
| Mean pairwise identity | 67.37 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -21.52 |
| Energy contribution | -18.80 |
| Covariance contribution | -2.72 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.78 |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.998353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
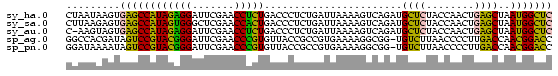
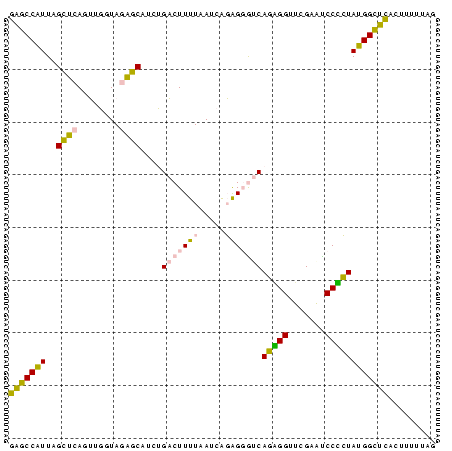
>sy_ha.0 1799678 81 - 2685015 CUAAUAAGUGAGCCAUAGAGGAUUCGAACCUCUGACCCUCUGAUUAAAAGUCAGAUGCUCUACCAACUGAGCUAAUGGCUC .........((((((((((((.......))))).....((((((.....)))))).((((........))))..))))))) ( -26.50) >sy_sa.0 1767392 81 - 2516575 CUUAAGAGUGAGCCAUAGUGGGCUCGAACCACUGACCCUCUGAUUAAAAGUCAGAUGCUCUACCAACUGAGCUAAUGGCUC .........((((((((((((.......))))).....((((((.....)))))).((((........))))..))))))) ( -27.60) >sy_au.0 2227973 80 + 2809422 C-AAGUAGUGAGCCAUAGAGGAUUCGAACCUCUGACCCUCUGAUUAAAAGUCAGAUGCUCUACCAACUGAGCUAAUGGCUC .-.......((((((((((((.......))))).....((((((.....)))))).((((........))))..))))))) ( -26.50) >sp_ag.0 1366783 80 + 2160267 GGCCACGAUAGUCCGUACGGGAUUCGAACCCGUGUUACCGCCGUGAAAAGGCGG-UGUCUUAACCCCUUGACCAACGGACC ..........(((((((((((.......)))))..(((((((.......)))))-)).................)))))). ( -28.40) >sp_pn.0 2033110 80 + 2038615 GGAUAAAAUAGUCCGUACGGGAUUCGAACCCGUGUUACCGCCGUGAAAAGGCGG-UGUCUUAACCCCUUGACCAACGGACC ..........(((((((((((.......)))))..(((((((.......)))))-)).................)))))). ( -28.40) >consensus CGAAAAAGUGAGCCAUAGAGGAUUCGAACCCCUGACCCUCUGAUUAAAAGUCAGAUGCUCUACCAACUGAGCUAAUGGCUC .........((((((((((((.......))))).......................((((........))))..))))))) (-21.52 = -18.80 + -2.72)
| Location | 1,799,678 – 1,799,759 |
|---|---|
| Length | 81 |
| Sequences | 5 |
| Columns | 81 |
| Reading direction | reverse |
| Mean pairwise identity | 67.37 |
| Mean single sequence MFE | -29.94 |
| Consensus MFE | -24.64 |
| Energy contribution | -22.64 |
| Covariance contribution | -2.00 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.997749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
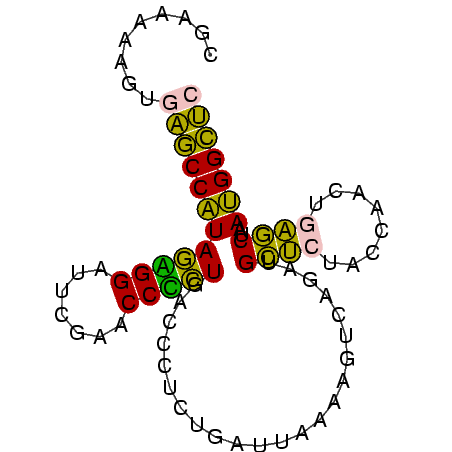
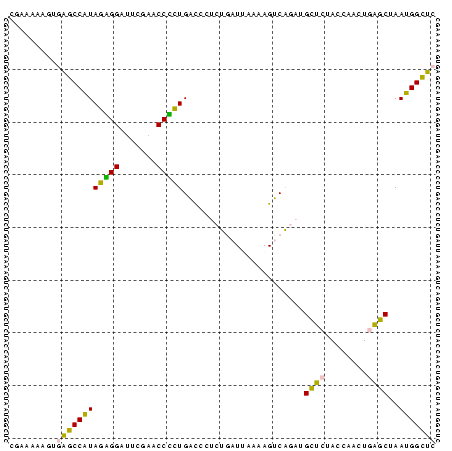
>sy_ha.0 1799678 81 - 2685015 GAGCCAUUAGCUCAGUUGGUAGAGCAUCUGACUUUUAAUCAGAGGGUCAGAGGUUCGAAUCCUCUAUGGCUCACUUAUUAG (((((((......((..((..((((.((((((((((.....)))))))))).))))....)).)))))))))......... ( -28.70) >sy_sa.0 1767392 81 - 2516575 GAGCCAUUAGCUCAGUUGGUAGAGCAUCUGACUUUUAAUCAGAGGGUCAGUGGUUCGAGCCCACUAUGGCUCACUCUUAAG ((((((...((((........))))..(((((((((.....)))))))))))))))(((((......)))))......... ( -31.50) >sy_au.0 2227973 80 + 2809422 GAGCCAUUAGCUCAGUUGGUAGAGCAUCUGACUUUUAAUCAGAGGGUCAGAGGUUCGAAUCCUCUAUGGCUCACUACUU-G (((((((......((..((..((((.((((((((((.....)))))))))).))))....)).))))))))).......-. ( -28.70) >sp_ag.0 1366783 80 + 2160267 GGUCCGUUGGUCAAGGGGUUAAGACA-CCGCCUUUUCACGGCGGUAACACGGGUUCGAAUCCCGUACGGACUAUCGUGGCC ((((((.(((((..((((((..((((-(((((.......)))))).......)))..)))))).....))))).)).)))) ( -32.01) >sp_pn.0 2033110 80 + 2038615 GGUCCGUUGGUCAAGGGGUUAAGACA-CCGCCUUUUCACGGCGGUAACACGGGUUCGAAUCCCGUACGGACUAUUUUAUCC (((((((..(((..........)))(-(((((.......))))))...(((((.......))))))))))))......... ( -28.80) >consensus GAGCCAUUAGCUCAGUUGGUAGAGCAUCUGACUUUUAAUCAGAGGGUCAGAGGUUCGAAUCCCCUAUGGCUCACUUUUUAG (((((((..((((........))))....(((((((.....)))))))(((((.......))))))))))))......... (-24.64 = -22.64 + -2.00) # Strand winner: reverse (0.98)
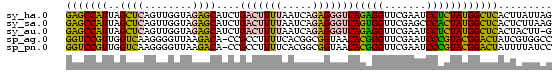
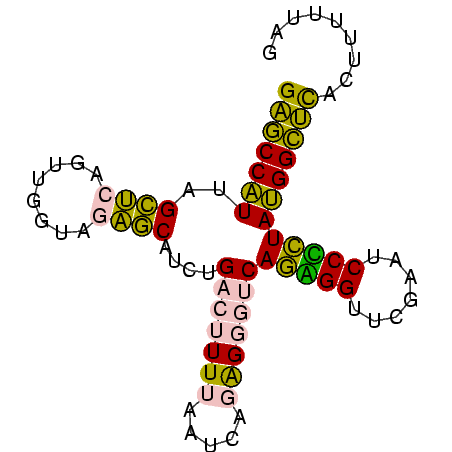
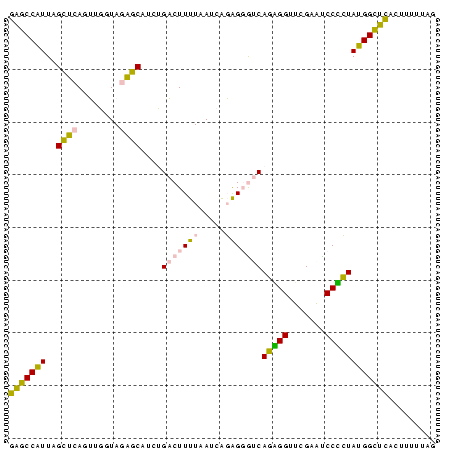
| Location | 1,799,678 – 1,799,759 |
|---|---|
| Length | 81 |
| Sequences | 5 |
| Columns | 81 |
| Reading direction | forward |
| Mean pairwise identity | 67.37 |
| Mean single sequence MFE | -27.48 |
| Consensus MFE | -21.52 |
| Energy contribution | -18.80 |
| Covariance contribution | -2.72 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.78 |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.998353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1799678 81 - 2685015/0-81 CUAAUAAGUGAGCCAUAGAGGAUUCGAACCUCUGACCCUCUGAUUAAAAGUCAGAUGCUCUACCAACUGAGCUAAUGGCUC .........((((((((((((.......))))).....((((((.....)))))).((((........))))..))))))) ( -26.50) >sy_sa.0 1767392 81 - 2516575/0-81 CUUAAGAGUGAGCCAUAGUGGGCUCGAACCACUGACCCUCUGAUUAAAAGUCAGAUGCUCUACCAACUGAGCUAAUGGCUC .........((((((((((((.......))))).....((((((.....)))))).((((........))))..))))))) ( -27.60) >sy_au.0 2227973 80 + 2809422/0-81 C-AAGUAGUGAGCCAUAGAGGAUUCGAACCUCUGACCCUCUGAUUAAAAGUCAGAUGCUCUACCAACUGAGCUAAUGGCUC .-.......((((((((((((.......))))).....((((((.....)))))).((((........))))..))))))) ( -26.50) >sp_ag.0 1366783 80 + 2160267/0-81 GGCCACGAUAGUCCGUACGGGAUUCGAACCCGUGUUACCGCCGUGAAAAGGCGG-UGUCUUAACCCCUUGACCAACGGACC ..........(((((((((((.......)))))..(((((((.......)))))-)).................)))))). ( -28.40) >sp_pn.0 2033110 80 + 2038615/0-81 GGAUAAAAUAGUCCGUACGGGAUUCGAACCCGUGUUACCGCCGUGAAAAGGCGG-UGUCUUAACCCCUUGACCAACGGACC ..........(((((((((((.......)))))..(((((((.......)))))-)).................)))))). ( -28.40) >consensus CGAAAAAGUGAGCCAUAGAGGAUUCGAACCCCUGACCCUCUGAUUAAAAGUCAGAUGCUCUACCAACUGAGCUAAUGGCUC .........((((((((((((.......))))).......................((((........))))..))))))) (-21.52 = -18.80 + -2.72)
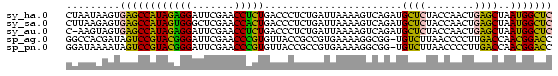
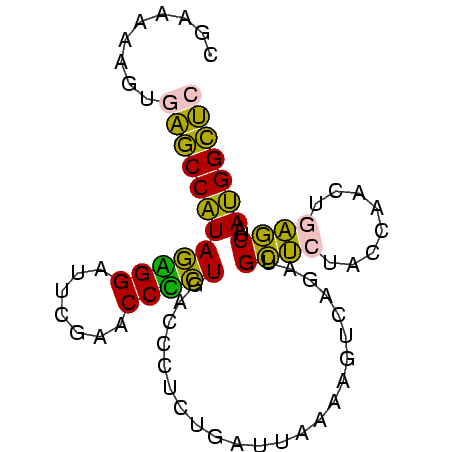
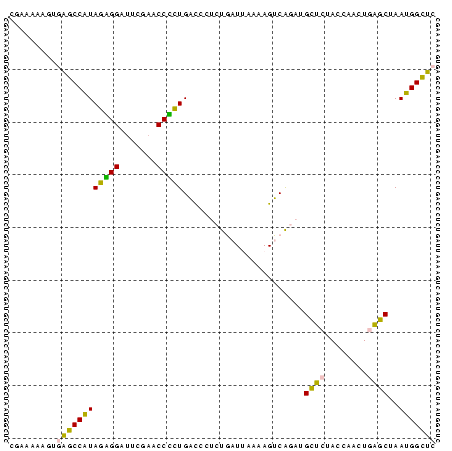
| Location | 1,799,678 – 1,799,759 |
|---|---|
| Length | 81 |
| Sequences | 5 |
| Columns | 81 |
| Reading direction | reverse |
| Mean pairwise identity | 67.37 |
| Mean single sequence MFE | -29.94 |
| Consensus MFE | -24.64 |
| Energy contribution | -22.64 |
| Covariance contribution | -2.00 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.997749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1799678 81 - 2685015/0-81 GAGCCAUUAGCUCAGUUGGUAGAGCAUCUGACUUUUAAUCAGAGGGUCAGAGGUUCGAAUCCUCUAUGGCUCACUUAUUAG (((((((......((..((..((((.((((((((((.....)))))))))).))))....)).)))))))))......... ( -28.70) >sy_sa.0 1767392 81 - 2516575/0-81 GAGCCAUUAGCUCAGUUGGUAGAGCAUCUGACUUUUAAUCAGAGGGUCAGUGGUUCGAGCCCACUAUGGCUCACUCUUAAG ((((((...((((........))))..(((((((((.....)))))))))))))))(((((......)))))......... ( -31.50) >sy_au.0 2227973 80 + 2809422/0-81 GAGCCAUUAGCUCAGUUGGUAGAGCAUCUGACUUUUAAUCAGAGGGUCAGAGGUUCGAAUCCUCUAUGGCUCACUACUU-G (((((((......((..((..((((.((((((((((.....)))))))))).))))....)).))))))))).......-. ( -28.70) >sp_ag.0 1366783 80 + 2160267/0-81 GGUCCGUUGGUCAAGGGGUUAAGACA-CCGCCUUUUCACGGCGGUAACACGGGUUCGAAUCCCGUACGGACUAUCGUGGCC ((((((.(((((..((((((..((((-(((((.......)))))).......)))..)))))).....))))).)).)))) ( -32.01) >sp_pn.0 2033110 80 + 2038615/0-81 GGUCCGUUGGUCAAGGGGUUAAGACA-CCGCCUUUUCACGGCGGUAACACGGGUUCGAAUCCCGUACGGACUAUUUUAUCC (((((((..(((..........)))(-(((((.......))))))...(((((.......))))))))))))......... ( -28.80) >consensus GAGCCAUUAGCUCAGUUGGUAGAGCAUCUGACUUUUAAUCAGAGGGUCAGAGGUUCGAAUCCCCUAUGGCUCACUUUUUAG (((((((..((((........))))....(((((((.....)))))))(((((.......))))))))))))......... (-24.64 = -22.64 + -2.00) # Strand winner: reverse (0.98)
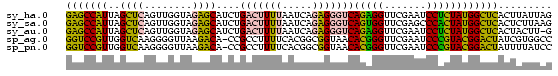
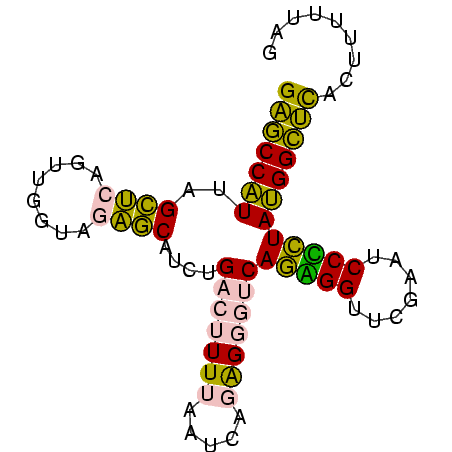
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:56:38 2006