
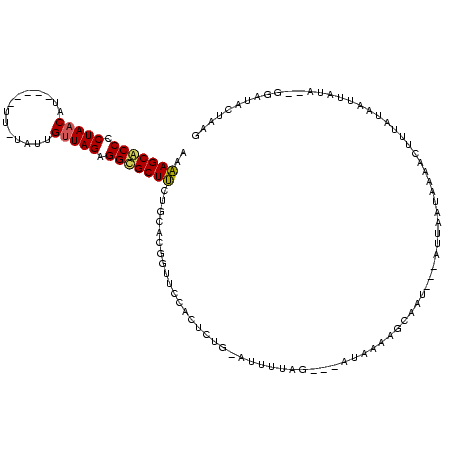
| Sequence ID | sy_sa.0 |
|---|---|
| Location | 2,287,061 – 2,287,178 |
| Length | 117 |
| Max. P | 0.999999 |

| Location | 2,287,061 – 2,287,164 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 58.17 |
| Mean single sequence MFE | -24.58 |
| Consensus MFE | -13.34 |
| Energy contribution | -13.34 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.54 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 2287061 103 + 2516575/0-120 AAAAGCACCCCUAACUU--------UAUUGUUAGAGGUGCUUUUGCACGGUUCCACUCUC-AUUUUA-----UAUAAGCAAUUGCCUAAAUCAAACUUUGUACUGAUU---GAUAUCAAG (((((((((.(((((..--------....))))).)))))))))...((((.........-..((((-----..(((....)))..))))...........)))).((---(....))). ( -22.65) >sy_au.0 2211593 119 - 2809422/0-120 AAAAGCACCACUAACAUACAAGUUGUAUUGUUAGAGGCGCUUCCGCACGGUUCCACUCUG-AAUUUAGCGAAUAACAUUAAUAAUAUUGCGGGCGCUUCCAAAUUAUCAAGGAAACUAAG ..((((.((.(((((((((.....))).)))))).)).))))(((((.(....).(.(((-....))).).................))))).................((....))... ( -24.50) >sy_ha.0 2480264 106 + 2685015/0-120 AUUAGCGCCCCUAACAC-----UUAAGAUGCUAGAGGCGCUACUGCACGGUUCCACUCUGUAUUCUAG---AUAAAUGCAA----GCUAGUAUGUCUGUAUAACUAUA--GGUUACUAGA ..(((((((.(((.(((-----....).)).))).))))))).((((.(....)..((((.....)))---)....)))).----.((((((...(((((....))))--)..)))))). ( -26.60) >consensus AAAAGCACCCCUAACAU_____UU_UAUUGUUAGAGGCGCUUCUGCACGGUUCCACUCUG_AUUUUAG___AUAAAAGCAAU___AUUAAUAAAACUUUAUAAUUAUA__GGAUACUAAG ..(((((((.(((((..............))))).))))))).............................................................................. (-13.34 = -13.34 + 0.00)

| Location | 2,287,061 – 2,287,164 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 58.17 |
| Mean single sequence MFE | -34.77 |
| Consensus MFE | -20.51 |
| Energy contribution | -19.74 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.84 |
| Structure conservation index | 0.59 |
| SVM decision value | 6.55 |
| SVM RNA-class probability | 0.999999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 2287061 103 + 2516575/0-120 CUUGAUAUC---AAUCAGUACAAAGUUUGAUUUAGGCAAUUGCUUAUA-----UAAAAU-GAGAGUGGAACCGUGCAAAAGCACCUCUAACAAUA--------AAGUUAGGGGUGCUUUU ..((((...---.))))((((...((((....(((((....)))))..-----..))))-......(....)))))(((((((((((((((....--------..))))))))))))))) ( -32.20) >sy_au.0 2211593 119 - 2809422/0-120 CUUAGUUUCCUUGAUAAUUUGGAAGCGCCCGCAAUAUUAUUAAUGUUAUUCGCUAAAUU-CAGAGUGGAACCGUGCGGAAGCGCCUCUAACAAUACAACUUGUAUGUUAGUGGUGCUUUU ....((((((..........))))))..(((((((((.....))))).((((((.....-...)))))).....))))(((((((.((((((.(((.....))))))))).))))))).. ( -38.20) >sy_ha.0 2480264 106 + 2685015/0-120 UCUAGUAACC--UAUAGUUAUACAGACAUACUAGC----UUGCAUUUAU---CUAGAAUACAGAGUGGAACCGUGCAGUAGCGCCUCUAGCAUCUUAA-----GUGUUAGGGGCGCUAAU .((((((..(--(.((....)).))...)))))).----((((((...(---(((..........))))...))))))(((((((((((((((.....-----))))))))))))))).. ( -33.90) >consensus CUUAGUAUCC__AAUAAUUACAAAGACACACUAAC___AUUGCUUUUAU___CUAAAAU_CAGAGUGGAACCGUGCAGAAGCGCCUCUAACAAUA_AA_____AUGUUAGGGGUGCUUUU ..............................................................(..((....))..)(((((((((((((((..............))))))))))))))) (-20.51 = -19.74 + -0.77) # Strand winner: reverse (1.00)


| Location | 2,287,061 – 2,287,178 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 63.97 |
| Mean single sequence MFE | -21.63 |
| Consensus MFE | -18.83 |
| Energy contribution | -16.41 |
| Covariance contribution | -2.43 |
| Combinations/Pair | 1.58 |
| Mean z-score | -0.12 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 2287061 117 + 2516575/160-280 AACCUAUGAUUAACAUUGAAAUCAACAAAGGAUUAGUAUGUGA---UGGACGGUGUCAAGAGAGUGUGCGGUAGCUGUGAGCACAUCACUUAAAUUACAGAAUGCACCUUUGUGCAAAUA ......(((((........)))))(((((((.......(((((---(....((((........((((((...........))))))))))...)))))).......)))))))....... ( -24.24) >sy_au.0 2211593 116 - 2809422/160-280 -AUGAAUAAAGUAUUUUAUAUAAAACGAAGGAUUAGUAAUUAAAUUUAUACGAUGC--AGAGAGUGUACGGUUGCUGUGAGUACAACGUAGAAAUUAAUGAAUGCACCUUCGUA-AAAUG -.......................(((((((....((((......))))....(((--(...(((.((((..(((.....)))...))))...)))......))))))))))).-..... ( -18.00) >sy_ha.0 2480264 116 + 2685015/160-280 AUUUUGGUAUGAGUAUUUAAA--GGCGAAGGAUUAGUAAAUAUUGGACUAUGAUAUC-AGAGAGUGUACGGAUGCUGUGAGUACGCCAUAGACGAUAUUGAAUGCACCUUUGUA-UAAUC .....................--.(((((((.......(((((((..(((((.....-.....((((((...........))))))))))).))))))).......))))))).-..... ( -22.64) >consensus AAUCUAUAAUGAAUAUUAAAA__AACGAAGGAUUAGUAAAUAA___UAUACGAUGUC_AGAGAGUGUACGGUUGCUGUGAGUACAACAUAGAAAUUAAUGAAUGCACCUUUGUA_AAAUA ........................(((((((.......(((((...((((((...........((((((...........))))))))))))..))))).......)))))))....... (-18.83 = -16.41 + -2.43) # Strand winner: reverse (0.98)


Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:56:35 2006