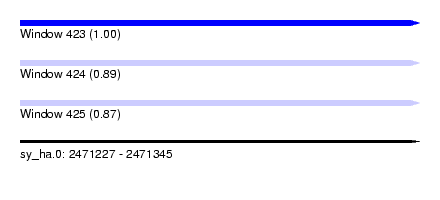
| Sequence ID | sy_ha.0 |
|---|---|
| Location | 2,471,227 – 2,471,345 |
| Length | 118 |
| Max. P | 0.998728 |

| Location | 2,471,227 – 2,471,345 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.14 |
| Mean single sequence MFE | -29.14 |
| Consensus MFE | -19.61 |
| Energy contribution | -18.73 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.67 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
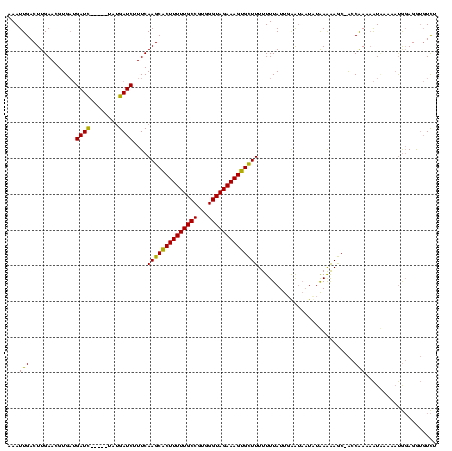
>sy_ha.0 2471227 118 + 2685015/0-120 AAAUUGACUUGAACGUG-UGAUCAUAAAAAUGAUCUUUCAAGCGCUUUUUGCCGUGGGUAGAAAGUGCUUUUUUUAUUGAGAAAUUUAAAAAGC-ACCAAAAAUAAAAAUGGAGGUGUCU (((((..((..(..(..-.((((((....))))))...)((((((((((((((...))))))))))))))......)..)).))))).....((-(((...............))))).. ( -30.26) >sy_au.0 2204346 113 - 2809422/0-120 AAAUUGACUUGAACGUGAUGAUC-----UAUGAUCUUUCAAGCACUUUUUGCCGUGGGUAGAAAGUGCUUUUUUUAUU-AAUUUUAAAAAAAGC-ACCAAAAAUUUAAAUGGAGGUGUCU ...((((...((.((((......-----)))).))..))))((((((((((((...))))))))))))((((((((..-.....))))))))((-(((...............))))).. ( -26.76) >sy_sa.0 2279440 115 + 2516575/0-120 AAAUUGACUUGAACGUGAAGAUU-----UUAGAUCUUUCAAGCACUUUUUGCCGUGGGUAGAAAGUGUUUUUUUUAUUGGAUAAUCUGAGGAACUGACGAACGUCAAUCCACAGAACUUC ................((((...-----((((((..(((((((((((((((((...))))))))))))........)))))..))))))(((..((((....)))).)))......)))) ( -30.40) >consensus AAAUUGACUUGAACGUGAUGAUC_____UAUGAUCUUUCAAGCACUUUUUGCCGUGGGUAGAAAGUGCUUUUUUUAUUGAAUAAUAUAAAAAGC_ACCAAAAAUAAAAAUGGAGGUGUCU ...(((.............((((........))))....((((((((((((((...))))))))))))))...........................))).................... (-19.61 = -18.73 + -0.88) # Strand winner: reverse (1.00)


| Location | 2,471,227 – 2,471,345 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.54 |
| Mean single sequence MFE | -22.17 |
| Consensus MFE | -18.63 |
| Energy contribution | -18.97 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
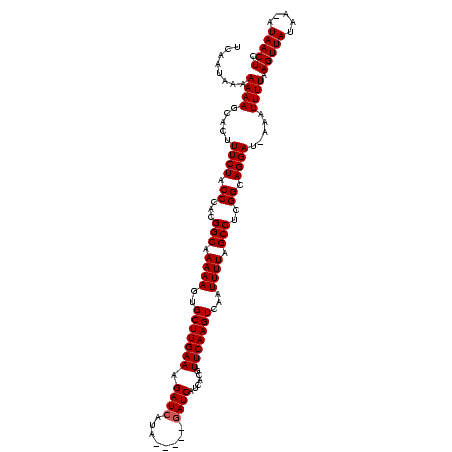
>sy_ha.0 2471227 118 + 2685015/40-160 UCAAUAAAAAAAGCACUUUCUACCCACGGCAAAAAGCGCUUGAAAGAUCAUUUUUAUGAUCA-CACGUUCAAGUCAAUUUUAGCCUCGGCAGGAU-AAUUUUUAAGUUAUACUAUAACUC .......(((((.....((((.((...(((.((((..(((((((.((((((....)))))).-....)))))))...)))).)))..)).)))).-..))))).((((((...)))))). ( -24.60) >sy_au.0 2204346 112 - 2809422/40-160 U-AAUAAAAAAAGCACUUUCUACCCACGGCAAAAAGUGCUUGAAAGAUCAUA-----GAUCAUCACGUUCAAGUCAAUUUUAGCCUCGGCAGGAU-AAAUUUUAAGUUAUAA-AUAACUC .-......((((.....((((.((...(((.((((.((((((((.((((...-----))))......)))))).)).)))).)))..)).)))).-...)))).(((((...-.))))). ( -19.20) >sy_sa.0 2279440 114 + 2516575/40-160 CCAAUAAAAAAAACACUUUCUACCCACGGCAAAAAGUGCUUGAAAGAUCUAA-----AAUCUUCACGUUCAAGUCAAUUUUAGCCUCGGUAGGAUCAAAUUUUAAGUUAUUA-CUAACUC .................(((((((...(((.((((.((((((((((((....-----.)))).....)))))).)).)))).)))..)))))))..........(((((...-.))))). ( -22.70) >consensus UCAAUAAAAAAAGCACUUUCUACCCACGGCAAAAAGUGCUUGAAAGAUCAUA_____GAUCAUCACGUUCAAGUCAAUUUUAGCCUCGGCAGGAU_AAAUUUUAAGUUAUAA_AUAACUC ........((((.....((((.((...(((.((((..(((((((.((((........))))......)))))))...)))).)))..)).)))).....)))).(((((.....))))). (-18.63 = -18.97 + 0.33)


| Location | 2,471,227 – 2,471,345 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.54 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -25.29 |
| Energy contribution | -24.63 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.868351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
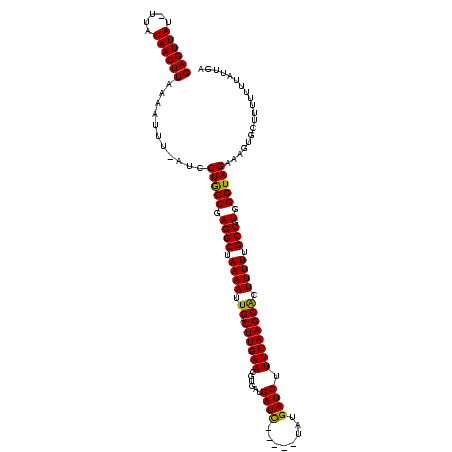
>sy_ha.0 2471227 118 + 2685015/40-160 GAGUUAUAGUAUAACUUAAAAAUU-AUCCUGCCGAGGCUAAAAUUGACUUGAACGUG-UGAUCAUAAAAAUGAUCUUUCAAGCGCUUUUUGCCGUGGGUAGAAAGUGCUUUUUUUAUUGA (((((((...))))))).......-...(((((.((((.((((.((.((((((....-.((((((....)))))).)))))))).)))).))).).)))))................... ( -28.50) >sy_au.0 2204346 112 - 2809422/40-160 GAGUUAU-UUAUAACUUAAAAUUU-AUCCUGCCGAGGCUAAAAUUGACUUGAACGUGAUGAUC-----UAUGAUCUUUCAAGCACUUUUUGCCGUGGGUAGAAAGUGCUUUUUUUAUU-A (((((((-(((.....)))).(((-(.(((....))).))))..)))))).....(((.((((-----...))))..)))(((((((((((((...))))))))))))).........-. ( -25.90) >sy_sa.0 2279440 114 + 2516575/40-160 GAGUUAG-UAAUAACUUAAAAUUUGAUCCUACCGAGGCUAAAAUUGACUUGAACGUGAAGAUU-----UUAGAUCUUUCAAGCACUUUUUGCCGUGGGUAGAAAGUGUUUUUUUUAUUGG ((((((.-...))))))..............((((((((((((((..(........)..))))-----)))).))))..((((((((((((((...))))))))))))))........)) ( -28.30) >consensus GAGUUAU_UUAUAACUUAAAAUUU_AUCCUGCCGAGGCUAAAAUUGACUUGAACGUGAUGAUC_____UAUGAUCUUUCAAGCACUUUUUGCCGUGGGUAGAAAGUGCUUUUUUUAUUGA ((((((.....))))))...........(((((.((((.((((.((.((((((......((((........)))).)))))))).)))).))).).)))))................... (-25.29 = -24.63 + -0.66) # Strand winner: reverse (1.00)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:56:28 2006