
| Sequence ID | sy_ha.0 |
|---|---|
| Location | 1,217,076 – 1,790,077 |
| Length | 573001 |
| Max. P | 0.999998 |

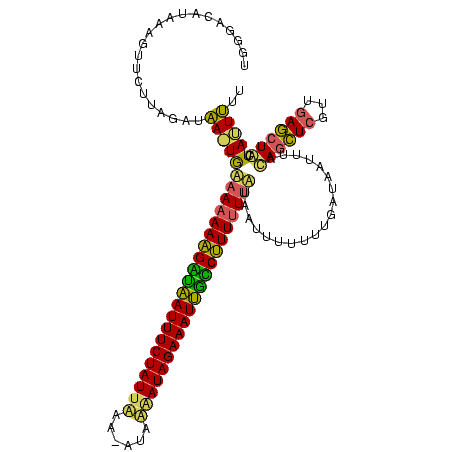
| Location | 1,789,962 – 1,790,077 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 63.40 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -24.69 |
| Energy contribution | -23.60 |
| Covariance contribution | -1.09 |
| Combinations/Pair | 1.44 |
| Mean z-score | -4.52 |
| Structure conservation index | 0.83 |
| SVM decision value | 6.49 |
| SVM RNA-class probability | 0.999998 |
| Prediction | RNA |
| WARNING | Sequence 1: Base composition out of range. |
| WARNING | Sequence 3: Base composition out of range. |
Download alignment: ClustalW | MAF
>sy_ha.0 1789962 115 - 2685015 UGGGACAUAAAGUUCUUGGAUAAGUGAAAAAAAGACAAUUUCUAUUGAA-AUAAUAUAGAAAUUGUCUUUUUUAUAAAUUUUUUGAUUAUUUUCAGCUCGUUGAGCUACUACUUUU .(((((.....))))).....(((((.((((((((((((((((((....-.....)))))))))))))))))).....................(((((...)))))..))))).. ( -28.30) >sy_au.0 1452647 115 + 2809422 AGUUCCUUAAAGUUCUUAGGCAA-UGUAAAAAAGCUGAUUUCUAUUAUUUAUUUGAUAGAAAUCAGCUUUUUUGAUAUGUAUUUUAUAAUGUACAGCUCGUUGAGCUGCUAUUUUC ..........(((((...(((..-..(((((((((((((((((((((......)))))))))))))))))))))...(((((........))))))))....)))))......... ( -36.90) >sy_sa.0 2167484 115 + 2516575 UAGGUCAUAAACAUCUUAGAUUAAUAAAACAACGGUAAUUUCUAUUUAA-UUCAAAUAGAAAUUGCCGUUUUCUUCAUUAUUAUGAAAAUUGAUAAUUCACUAAACUAUUAUUGUU .........((((...(((...........(((((((((((((((((..-...)))))))))))))))))...(((((....)))))..................)))....)))) ( -24.20) >consensus UGGGACAUAAAGUUCUUAGAUAA_UGAAAAAAAGAUAAUUUCUAUUAAA_AUAAAAUAGAAAUUGCCUUUUUUAUAAUUUUUUUGAUAAUUUACAGCUCGUUGAGCUACUAUUUUU .....................((((((((((((((((((((((((((......)))))))))))))))))))))...................((((((...)))))).))))).. (-24.69 = -23.60 + -1.09)

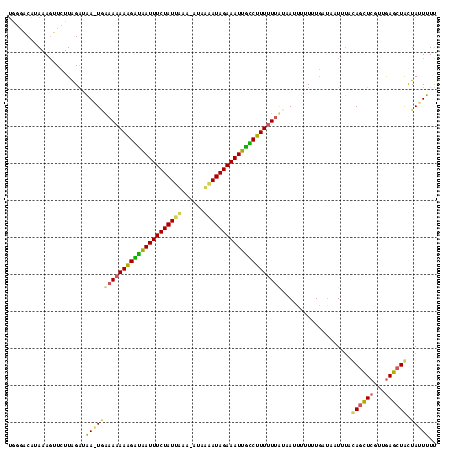
| Location | 1,789,962 – 1,790,077 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 63.40 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -24.90 |
| Energy contribution | -22.47 |
| Covariance contribution | -2.43 |
| Combinations/Pair | 1.48 |
| Mean z-score | -4.89 |
| Structure conservation index | 0.91 |
| SVM decision value | 5.06 |
| SVM RNA-class probability | 0.999971 |
| Prediction | RNA |
| WARNING | Sequence 1: Base composition out of range. |
| WARNING | Sequence 3: Base composition out of range. |
Download alignment: ClustalW | MAF
>sy_ha.0 1789962 115 - 2685015 AAAAGUAGUAGCUCAACGAGCUGAAAAUAAUCAAAAAAUUUAUAAAAAAGACAAUUUCUAUAUUAU-UUCAAUAGAAAUUGUCUUUUUUUCACUUAUCCAAGAACUUUAUGUCCCA .(((((..((((((...))))))....................((((((((((((((((((.....-....))))))))))))))))))..............)))))........ ( -26.20) >sy_au.0 1452647 115 + 2809422 GAAAAUAGCAGCUCAACGAGCUGUACAUUAUAAAAUACAUAUCAAAAAAGCUGAUUUCUAUCAAAUAAAUAAUAGAAAUCAGCUUUUUUACA-UUGCCUAAGAACUUUAAGGAACU .......(((((((...)))))))...................((((((((((((((((((..........))))))))))))))))))...-...(((..........))).... ( -30.70) >sy_sa.0 2167484 115 + 2516575 AACAAUAAUAGUUUAGUGAAUUAUCAAUUUUCAUAAUAAUGAAGAAAACGGCAAUUUCUAUUUGAA-UUAAAUAGAAAUUACCGUUGUUUUAUUAAUCUAAGAUGUUUAUGACCUA ((((....(((.((((((((.......(((((((....))))))).(((((.((((((((((((..-.)))))))))))).)))))..)))))))).)))...))))......... ( -25.60) >consensus AAAAAUAGUAGCUCAACGAGCUGUAAAUUAUCAAAAAAAUAAAAAAAAAGACAAUUUCUAUAUAAU_UUAAAUAGAAAUUACCUUUUUUUCA_UUAUCUAAGAACUUUAUGACCCA .......(((((((...)))))))...................((((((((((((((((((..........))))))))))))))))))........................... (-24.90 = -22.47 + -2.43) # Strand winner: forward (0.57)

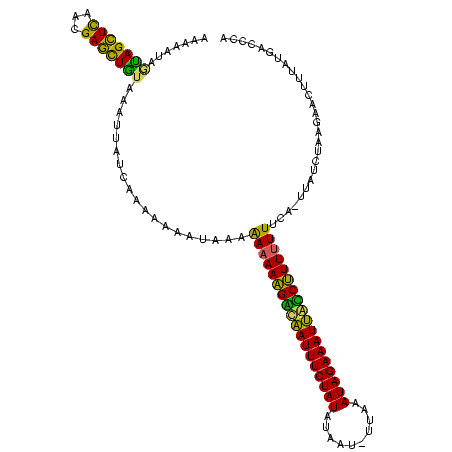
| Location | 1,217,076 – 1,217,121 |
|---|---|
| Length | 45 |
| Sequences | 3 |
| Columns | 45 |
| Reading direction | forward |
| Mean pairwise identity | 80.74 |
| Mean single sequence MFE | -21.50 |
| Consensus MFE | -21.48 |
| Energy contribution | -20.93 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.35 |
| Mean z-score | -7.28 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897427 |
| Prediction | RNA |
| WARNING | Sequence 1 too short. |
| WARNING | Sequence 2 too short. |
| WARNING | Sequence 3 too short. |
Download alignment: ClustalW | MAF
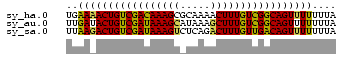
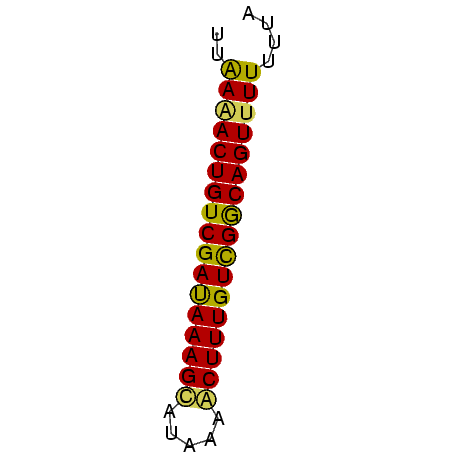
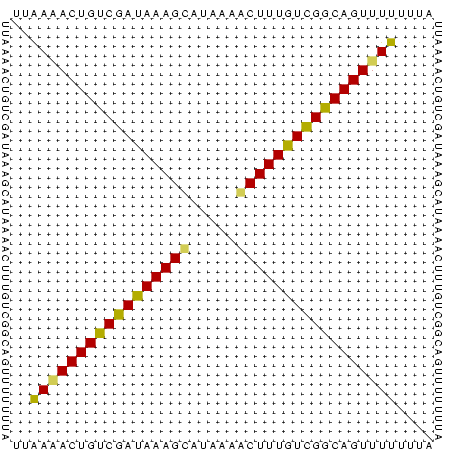
>sy_ha.0 1217076 45 + 2685015 UGAAAACUGUCGACAAAGCGCAAAACUUUGUCGGCAGUUUUUUUA .(((((((((((((((((.......)))))))))))))))))... ( -22.40) >sy_au.0 549602 45 + 2809422 UUGAUACUGUCGAUAAAGCAUAAAGCUUUGUCGGCAGUUUUUUUA .....((((((((((((((.....))))))))))))))....... ( -20.60) >sy_sa.0 2250489 45 + 2516575 UUAAGACUGUCGAUAAAGUCUCAGACUUUGUUGACAGUUUUUUUA ..((((((((((((((((((...)))))))))))))))))).... ( -21.50) >consensus UUAAAACUGUCGAUAAAGCAUAAAACUUUGUCGGCAGUUUUUUUA ..(((((((((((((((((.....))))))))))))))))).... (-21.48 = -20.93 + -0.54)
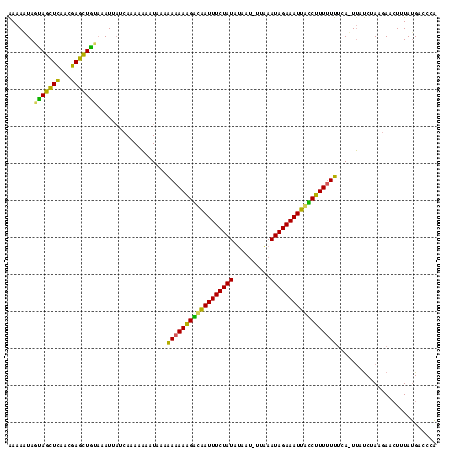
| Location | 1,217,076 – 1,217,121 |
|---|---|
| Length | 45 |
| Sequences | 3 |
| Columns | 45 |
| Reading direction | reverse |
| Mean pairwise identity | 80.74 |
| Mean single sequence MFE | -14.30 |
| Consensus MFE | -9.81 |
| Energy contribution | -9.70 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -4.79 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996779 |
| Prediction | RNA |
| WARNING | Sequence 1 too short. |
| WARNING | Sequence 2 too short. |
| WARNING | Sequence 3 too short. |
Download alignment: ClustalW | MAF
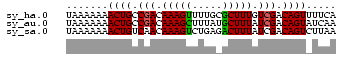
>sy_ha.0 1217076 45 + 2685015 UAAAAAAACUGCCGACAAAGUUUUGCGCUUUGUCGACAGUUUUCA ....(((((((.(((((((((.....))))))))).))))))).. ( -18.60) >sy_au.0 549602 45 + 2809422 UAAAAAAACUGCCGACAAAGCUUUAUGCUUUAUCGACAGUAUCAA .......((((.(((.(((((.....))))).))).))))..... ( -13.30) >sy_sa.0 2250489 45 + 2516575 UAAAAAAACUGUCAACAAAGUCUGAGACUUUAUCGACAGUCUUAA .......((((((...((((((...))))))...))))))..... ( -11.00) >consensus UAAAAAAACUGCCGACAAAGUUUUACGCUUUAUCGACAGUAUUAA .......((((.(((.(((((.....))))).))).))))..... ( -9.81 = -9.70 + -0.11) # Strand winner: forward (1.00)
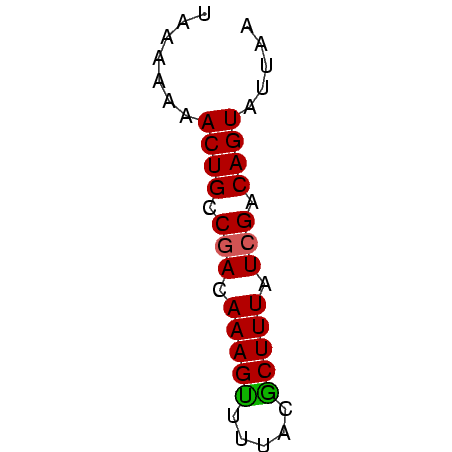
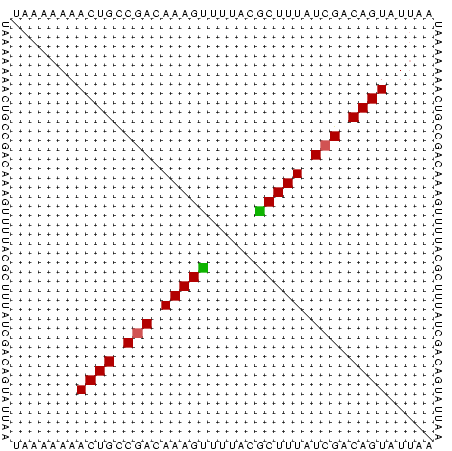
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:56:18 2006