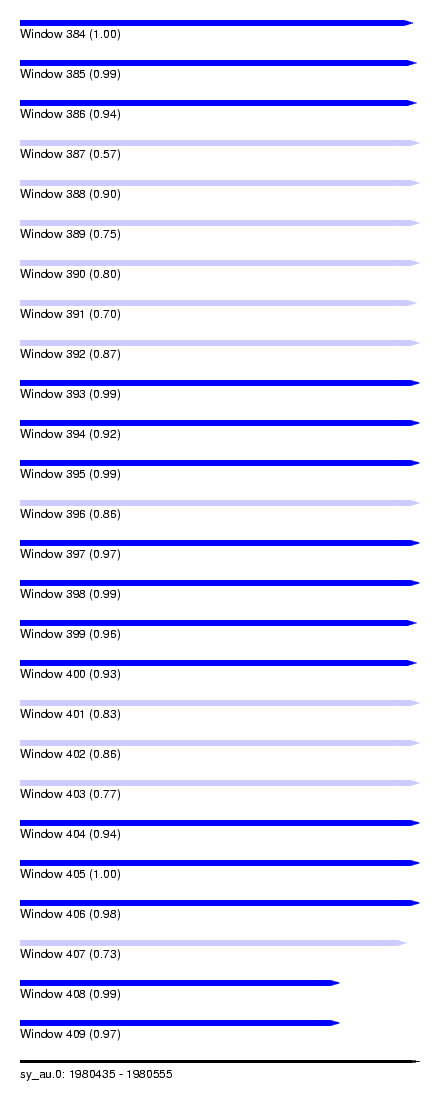
| Sequence ID | sy_au.0 |
|---|---|
| Location | 1,980,435 – 1,980,555 |
| Length | 120 |
| Max. P | 0.998936 |

| Location | 1,980,435 – 1,980,553 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.70 |
| Mean single sequence MFE | -36.98 |
| Consensus MFE | -23.20 |
| Energy contribution | -24.33 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.63 |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 118 + 2809422/0-120 AGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUUCUUCAGAAGAUGC-GGAAUAACG-UGACAUAUUGUAUUCAGUUUUGAAUGUUUGUU .((((((((((((....))))))))).....)))((((.....)))).(((((((((((((((((....))))))..-.(((((.((-........))))))).)))))))))))..... ( -40.20) >sy_sa.0 1555301 120 - 2516575/0-120 AGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUUCUUACGAAGAUGCAGGAAUAACAUUGACAUAUUGUAUUCAGUUUUGAAUGCUCAUU .((((((((((((....))))))))).....)))((((.....))))..((((((((((((((((....))))))....(((((.((.........))))))).))))))))))...... ( -38.90) >sy_ha.0 1580250 115 - 2685015/0-120 AGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUCGUAGAGAUGA----GGAAUAACG-UGACAUAUUGUAUUCAGUUUUGAAUGUUUAUU .((((((((((((....))))))))).....)))((((.....)))).(((((((((((..(((((....))))----)(((((.((-........))))))).)))))))))))..... ( -38.40) >sp_ag.0 1993425 93 - 2160267/0-120 AGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUA-----AGGAAACCUGC--------------------CAUUUGCGUCUUG--UUUAGUUUUGAGAGGUCUUG (((((((((((((....)))))))))........(((((((....)).)-----)))).))))..--------------------.....((.(((((--........))))).)).... ( -30.40) >consensus AGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUUCUU_AGAAGA____GGAAUAACA_UGACAUAUUGUAUUCAGUUUUGAAUGUUCAUU .((((((((((((....))))))))).....)))((((.....))))..((((((((((.............................................))))))))))...... (-23.20 = -24.33 + 1.13) # Strand winner: reverse (1.00)

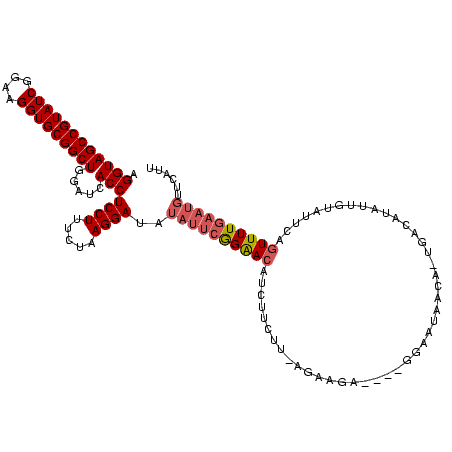
| Location | 1,980,435 – 1,980,554 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.43 |
| Mean single sequence MFE | -40.92 |
| Consensus MFE | -32.81 |
| Energy contribution | -34.00 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 119 + 2809422/40-160 CGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUUCUUCAGAAGAUGC-GG ..((((((((((..(..((((((.......))))))........(((((((((....))))))))))..)))))((((.....))))....)))))..(((((((....))))))).-.. ( -43.90) >sy_sa.0 1555301 120 - 2516575/40-160 CGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUUCUUACGAAGAUGCAGG ..((((((((((..(..((((((.......))))))........(((((((((....))))))))))..)))))((((.....))))....)))))..(((((((....))))))).... ( -43.60) >sy_ha.0 1580250 116 - 2685015/40-160 CGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUCGUAGAGAUGA----GG ..((((((((((..(..((((((.......))))))........(((((((((....))))))))))..)))))((((.....))))....)))))..((((((...)))))).----.. ( -41.90) >sp_ag.0 1993425 100 - 2160267/40-160 CGCCUAAGGUGGGAUAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUA-----AGGAAACCUGC--------------- ......(((((((((..((((((.......)))))).......((((((((((....)))))))))).))).))(((((((....)).)-----)))).))))..--------------- ( -34.30) >consensus CGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUAUUCGGAACAUCUUCUU_AGAAGA____GG ....(((((((...(....((..(((((((.((....))....((((((((((....))))))))))..))))).))..))....((.....)))...)))))))............... (-32.81 = -34.00 + 1.19) # Strand winner: reverse (1.00)

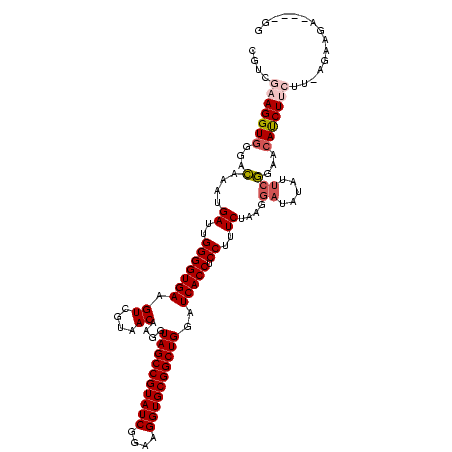
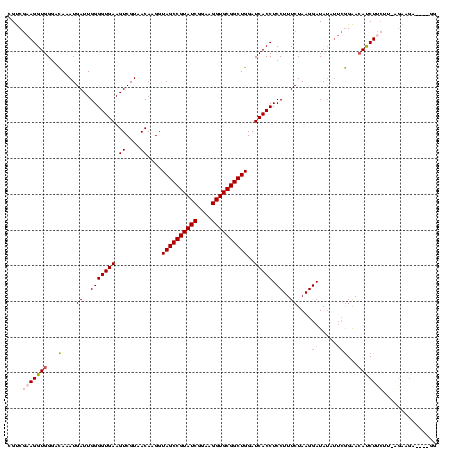
| Location | 1,980,435 – 1,980,554 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.26 |
| Mean single sequence MFE | -43.35 |
| Consensus MFE | -42.35 |
| Energy contribution | -41.98 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 119 + 2809422/80-200 AACACCCGAAGCCGGUGGAGUAACCUUUUA-GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUU ..(((((((...((((..(((..((.....-)).))).)))))))..))))............(((((((.((....))....((((((((((....))))))))))..))))))).... ( -42.60) >sy_sa.0 1555301 120 - 2516575/80-200 AACACCCGAAGCCGGUGGAGUAACCAUUUAUGGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUU ..(((((((...((((..(((..(((....))).))).)))))))..))))............(((((((.((....))....((((((((((....))))))))))..))))))).... ( -43.30) >sy_ha.0 1580250 118 - 2685015/80-200 AACACCCGAAGCCGGUGGAGUAACCAUUU--GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUU ..((((.......)))).......(((((--(...((.(((......))).)).))))))...(((((((.((....))....((((((((((....))))))))))..))))))).... ( -43.20) >sp_ag.0 1993425 119 - 2160267/80-200 AACACCCGAAGUCGGUGAGGUAACCUUUUA-GGAGCCAGCCGCCUAAGGUGGGAUAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUU ..((((...((.((((..(((..((.....-)).))).)))).))..))))............(((((((.((....))....((((((((((....))))))))))..))))))).... ( -44.30) >consensus AACACCCGAAGCCGGUGGAGUAACCAUUUA_GGAGCUAGCCGUCGAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUU ..((((....(.((((..(((..((......)).))).)))).)...))))............(((((((.((....))....((((((((((....))))))))))..))))))).... (-42.35 = -41.98 + -0.37) # Strand winner: reverse (1.00)

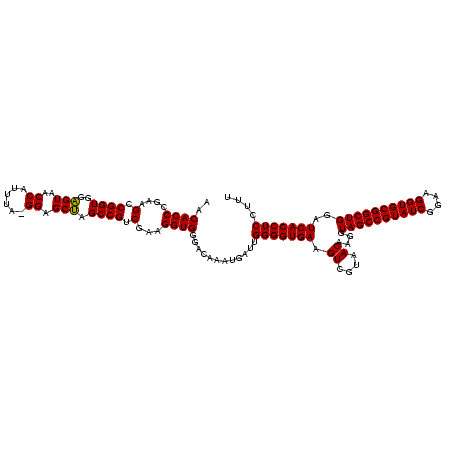
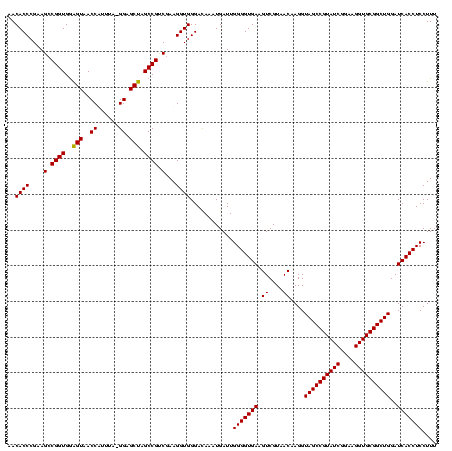
| Location | 1,980,435 – 1,980,555 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.92 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -33.74 |
| Energy contribution | -30.18 |
| Covariance contribution | -3.56 |
| Combinations/Pair | 1.38 |
| Mean z-score | -0.93 |
| Structure conservation index | 1.05 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 120 + 2809422/280-400 ACACACGUGCUACAAUGGACAAUACAAAGGGCAGCGAAACCGUGAGGUCAAGCAAAUCCCAUAAAGUUGUUCUCAGUUCGGAUUGUAGUCUGCAACUCGACUACAUGAAGCUGGAAUCGC ........((......(((((((.....(((..((...(((....)))...))....))).....)))))))((((((.....(((((((........)))))))...))))))....)) ( -31.50) >sy_sa.0 1555301 120 - 2516575/280-400 ACACACGUGCUACAAUGGACAAUACAAAGGGCAGCUAAACCGCGAGGUCAUGCAAAUCCCAUAAAGUUGUUCUCAGUUCGGAUUGUAGUCUGCAACUCGACUACAUGAAGCUGGAAUCGC ........((......(((((((.....(((..((...(((....)))...))....))).....)))))))((((((.....(((((((........)))))))...))))))....)) ( -30.60) >sy_ha.0 1580250 120 - 2685015/280-400 ACACACGUGCUACAAUGGACAAUACAAAGGGCAGCGAAAUCGCGAGGUCAAGCAAAUCCCAUAAAGUUGUUCUCAGUUCGGAUUGUAGUCUGCAACUCGACUACAUGAAGCUGGAAUCGC ........((......(((((((.....(((..((...(((....)))...))....))).....)))))))((((((.....(((((((........)))))))...))))))....)) ( -28.40) >sp_ag.0 1993425 120 - 2160267/280-400 ACACACGUGCUACAAUGGUUGGUACAACGAGUCGCAAGCCGGUGACGGCAAGCUAAUCUCUUAAAGCCAAUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAGUCGGAAUCGC ........((......(((((((.....(((..((..((((....))))..))....))).....)))))))...((((.((((((((((........)))))))....))).)))).)) ( -38.60) >consensus ACACACGUGCUACAAUGGACAAUACAAAGGGCAGCGAAACCGCGAGGUCAAGCAAAUCCCAUAAAGUUGUUCUCAGUUCGGAUUGUAGUCUGCAACUCGACUACAUGAAGCUGGAAUCGC ........((......(((((((.....(((..((...(((....)))...))....))).....)))))))....(((((..(((((((........))))))).....)))))...)) (-33.74 = -30.18 + -3.56) # Strand winner: reverse (0.60)

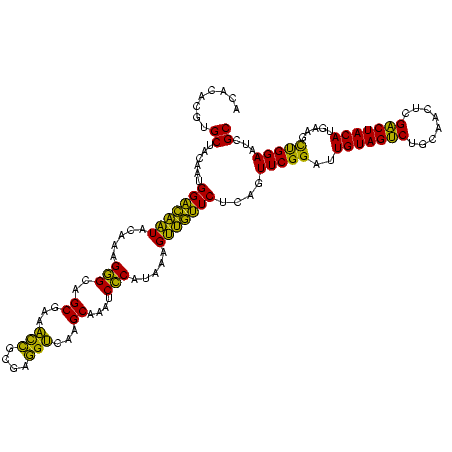
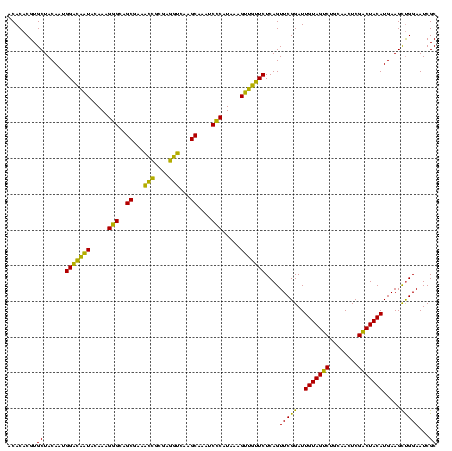
| Location | 1,980,435 – 1,980,555 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -37.03 |
| Consensus MFE | -36.71 |
| Energy contribution | -36.53 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 120 + 2809422/400-520 AGCCCAAAUCAUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCAACUUAGAGUGCCCAACUUAAUGAUGGCAACUAAGCUUAAGGGUUGCGCUC (((((..(((((..((((.((((((......))))))))))..........(((.....)))((((.((......)).)))).......)))))(((......)))...)))))...... ( -36.60) >sy_sa.0 1555301 120 - 2516575/400-520 AGCCCAAAUCAUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCAACCUAGAGUGCCCAACUUAAUGAUGGCAACUAAGCUUAAGGGUUGCGCUC (((((..(((((..((((.((((((......))))))))))..........(((.....)))((((.((......)).)))).......)))))(((......)))...)))))...... ( -36.60) >sy_ha.0 1580250 120 - 2685015/400-520 AGCCCAAAUCAUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCAACUUAGAGUGCCCAACUUAAUGAUGGCAACUAAACUUAAGGGUUGCGCUC (((....(((((..((((.((((((......))))))))))..........(((.....)))((((.((......)).)))).......))))).((((((.........))))))))). ( -36.20) >sp_ag.0 1993425 120 - 2160267/400-520 AGCCCAGGUCAUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUAUUACCGGCAGUCUCGCUAGAGUGCCCAACUUAAUGAUGGCAACUAACAAUAGGGGUUGCGCUC (((..((((.....((((.((((((......))))))))))))))..(((((((.....))))).))....))).((((((....((((.((..(....)...)).))))....)))))) ( -38.70) >consensus AGCCCAAAUCAUAAGGGGCAUGAUGAUUUGACGUCAUCCCCACCUUCCUCCGGUUUGUCACCGGCAGUCAACCUAGAGUGCCCAACUUAAUGAUGGCAACUAAGCUUAAGGGUUGCGCUC (((....(((((..((((.((((((......))))))))))..........(((.....)))((((.((......)).)))).......))))).((((((.........))))))))). (-36.71 = -36.53 + -0.19)



| Location | 1,980,435 – 1,980,555 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -40.15 |
| Consensus MFE | -39.30 |
| Energy contribution | -37.80 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 120 + 2809422/400-520 GAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAAGUUGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAUGAUUUGGGCU .(((.(((.((((.(((((((.((((..(.....)...))))..)))))))..((.(((((.....))))).)))))).((((((((.(......).)))).)))).......))).))) ( -40.90) >sy_sa.0 1555301 120 - 2516575/400-520 GAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAGGUUGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAUGAUUUGGGCU .(((.(((.((((.(((((((.((((..(.....)...))))..)))))))..((.(((((.....))))).)))))).((((((((.(......).)))).)))).......))).))) ( -40.60) >sy_ha.0 1580250 120 - 2685015/400-520 GAGCGCAACCCUUAAGUUUAGUUGCCAUCAUUAAGUUGGGCACUCUAAGUUGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAUGAUUUGGGCU .(((.(((.((((..((((.((..((..((......))((((.((......)).))))))..))))))..)))).(((.(((.((((.(......).)))).)))))).....))).))) ( -39.90) >sp_ag.0 1993425 120 - 2160267/400-520 GAGCGCAACCCCUAUUGUUAGUUGCCAUCAUUAAGUUGGGCACUCUAGCGAGACUGCCGGUAAUAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAUGACCUGGGCU .(((.((((((((.(((((((.((((..(.....)...))))..))))))).....(((((.....))))))))..)))((((((((.(......).)))).))))........)).))) ( -39.20) >consensus GAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAAGUUGACUGCCGGUGACAAACCGGAGGAAGGUGGGGAUGACGUCAAAUCAUCAUGCCCCUUAUGAUUUGGGCU .(((.(((((....(((((((.((((..(.....)...))))..)))))))..((.(((((.....))))).))..)))((((((((.(......).)))).))))........)).))) (-39.30 = -37.80 + -1.50) # Strand winner: reverse (0.98)

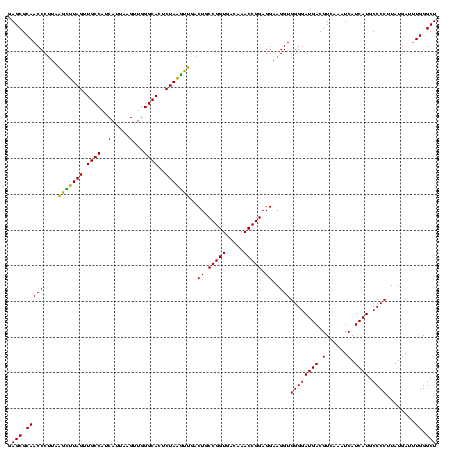
| Location | 1,980,435 – 1,980,555 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -42.95 |
| Consensus MFE | -42.53 |
| Energy contribution | -41.03 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.800094 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 120 + 2809422/440-560 GUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAAGUUGACUGCCGGUGACAAACCGGAGGAAGGUG ....(((((((((....)))((.(((......))))).)))))).....((((.(((((((.((((..(.....)...))))..)))))))..((.(((((.....))))).)))))).. ( -43.50) >sy_sa.0 1555301 120 - 2516575/440-560 GUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAGGUUGACUGCCGGUGACAAACCGGAGGAAGGUG ....(((((((((....)))((.(((......))))).)))))).....((((.(((((((.((((..(.....)...))))..)))))))..((.(((((.....))))).)))))).. ( -43.20) >sy_ha.0 1580250 120 - 2685015/440-560 GUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUAAGUUUAGUUGCCAUCAUUAAGUUGGGCACUCUAAGUUGACUGCCGGUGACAAACCGGAGGAAGGUG ....(((((((((....)))((.(((......))))).))))))((...((((..((((.((..((..((......))((((.((......)).))))))..))))))..))))...)). ( -42.60) >sp_ag.0 1993425 120 - 2160267/440-560 GUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCCUAUUGUUAGUUGCCAUCAUUAAGUUGGGCACUCUAGCGAGACUGCCGGUAAUAAACCGGAGGAAGGUG ....(((((((((....)))((.(((......))))).))))))...((((((.(((((((.((((..(.....)...))))..))))))).....(((((.....))))))))..))). ( -42.50) >consensus GUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCGCAACCCUUAAGCUUAGUUGCCAUCAUUAAGUUGGGCACUCUAAGUUGACUGCCGGUGACAAACCGGAGGAAGGUG ....(((((((((....)))((.(((......))))).))))))...(((....(((((((.((((..(.....)...))))..)))))))..((.(((((.....))))).))..))). (-42.53 = -41.03 + -1.50) # Strand winner: reverse (1.00)


| Location | 1,980,435 – 1,980,554 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.47 |
| Mean single sequence MFE | -37.67 |
| Consensus MFE | -32.22 |
| Energy contribution | -31.98 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 119 + 2809422/560-680 GCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUUACCAAAUCUUGACAUCCUU-UGACAACUCUAGAGAUAGAGCUUUCCCCUUCGGGGGACAAAGUGACAGGUGGUGCAUGGUUGUC .((((((((((..((((........)))))))))..((((...((((...(((((-((....(((((....)))))...(((((....)))))))))).)))))))))))))))...... ( -37.90) >sy_sa.0 1555301 119 - 2516575/560-680 GCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUUACCAAAUCUUGACAUCCUU-UGAAAACUCUAGAGAUAGAGCCUUCCCCUUCGGGGGACAAAGUGACAGGUGGUGCAUGGUUGUC .((((((((((..((((........)))))))))..((((...((((...(((((-((....(((((....)))))...(((((....)))))))))).)))))))))))))))...... ( -37.90) >sy_ha.0 1580250 119 - 2685015/560-680 GCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUUACCAAAUCUUGACAUCCUU-UGACAACUCUAGAGAUAGAGCCUUCCCCUUCGGGGGACAAAGUGACAGGUGGUGCAUGGUUGUC .((((((((((..((((........)))))))))..((((...((((...(((((-((....(((((....)))))...(((((....)))))))))).)))))))))))))))...... ( -37.90) >sp_ag.0 1993425 117 - 2160267/560-680 GCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUUACCAGGUCUUGACAUCCUUCUGACCGGCCUAGAGAUAGG--CUUUCUCUUCGGAGCA-GAAGUGACAGGUGGUGCAUGGUUGUC .((((((((((..((((........)))))))))..((((...((((...((((((((...((((((....))))--))..(((....)))))-)))).)))))))))))))))...... ( -37.00) >consensus GCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUUACCAAAUCUUGACAUCCUU_UGACAACUCUAGAGAUAGAGCCUUCCCCUUCGGGGGACAAAGUGACAGGUGGUGCAUGGUUGUC .((((((((((..((((........)))))))))..((((...((((.(((...........(((((....)))))...(((((....)))))....))).)))))))))))))...... (-32.22 = -31.98 + -0.25) # Strand winner: reverse (0.98)


| Location | 1,980,435 – 1,980,555 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -43.22 |
| Consensus MFE | -41.01 |
| Energy contribution | -40.57 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.873721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 120 + 2809422/680-800 UCCACCGCUUGUGCGGGUCCCCGUCAAUUCCUUUGAGUUUCAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUU ....(((((((.((((....))))..........((((..(.(((....))).).))))..)))))))((((((....((....))...)))))).(((((.(....))))))....... ( -44.40) >sy_sa.0 1555301 120 - 2516575/680-800 UCCACCGCUUGUGCGGGUCCCCGUCAAUUCCUUUGAGUUUCAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUU ....(((((((.((((....))))..........((((..(.(((....))).).))))..)))))))((((((....((....))...)))))).(((((.(....))))))....... ( -44.40) >sy_ha.0 1580250 120 - 2685015/680-800 UCCACCGCUUGUGCGGGUCCCCGUCAAUUCCUUUGAGUUUCAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUU ....(((((((.((((....))))..........((((..(.(((....))).).))))..)))))))((((((....((....))...)))))).(((((.(....))))))....... ( -44.40) >sp_ag.0 1993425 120 - 2160267/680-800 UCCACCGCUUGUGCGGGCCCCCGUCAAUUCCUUUGAGUUUCAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCGGCACUAAGCCCCGGAAAGGGCCUAACACCU .....(((....)))((((((((...........((((..(.(((....))).).))))....(((..((((((....((....))...))))))..))).)))...)))))........ ( -39.70) >consensus UCCACCGCUUGUGCGGGUCCCCGUCAAUUCCUUUGAGUUUCAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUU ....(((((((.((((....))))..........((((..(.(((....))).).))))..)))))))((((((....((....))...)))))).(((((.(....))))))....... (-41.01 = -40.57 + -0.44)

| Location | 1,980,435 – 1,980,555 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -50.45 |
| Consensus MFE | -51.45 |
| Energy contribution | -49.95 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.83 |
| Structure conservation index | 1.02 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 120 + 2809422/680-800 AAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCGGUGGA .((..(((((((((....)))))))))..))...((....))........((((((((...((((.(((((....)))))..))))..)))......))))).(((((....)))).).. ( -49.40) >sy_sa.0 1555301 120 - 2516575/680-800 AAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCGGUGGA .((..(((((((((....)))))))))..))...((....))........((((((((...((((.(((((....)))))..))))..)))......))))).(((((....)))).).. ( -49.40) >sy_ha.0 1580250 120 - 2685015/680-800 AAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCGGUGGA .((..(((((((((....)))))))))..))...((....))........((((((((...((((.(((((....)))))..))))..)))......))))).(((((....)))).).. ( -49.40) >sp_ag.0 1993425 120 - 2160267/680-800 AGGUGUUAGGCCCUUUCCGGGGCUUAGUGCCGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGA .((..(((((((((....)))))))))..))...((....)).....((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))).... ( -53.60) >consensus AAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCGGUGGA .((..(((((((((....)))))))))..))...((....))........((((((((...((((.(((((....)))))..))))..)))......))))).(((((....)))).).. (-51.45 = -49.95 + -1.50) # Strand winner: reverse (0.98)

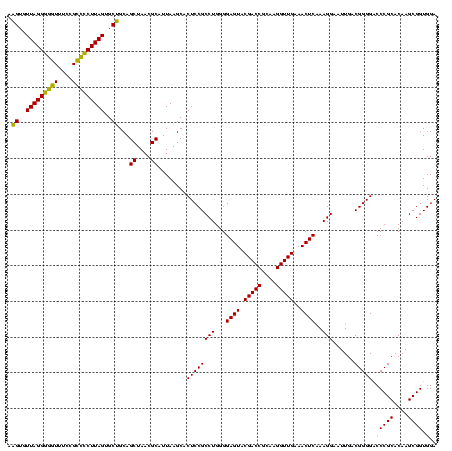
| Location | 1,980,435 – 1,980,555 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -44.02 |
| Consensus MFE | -41.88 |
| Energy contribution | -40.75 |
| Covariance contribution | -1.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 120 + 2809422/720-840 CAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAA ..((((((.((((.(((.((..((((((((((((....(.((((((....)).....((((.(....))))))))).)..)))))))..)))))..)).)))))))..))))))...... ( -44.90) >sy_sa.0 1555301 120 - 2516575/720-840 CAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAA ..((((((.((((.(((.((..((((((((((((....(.((((((....)).....((((.(....))))))))).)..)))))))..)))))..)).)))))))..))))))...... ( -44.90) >sy_ha.0 1580250 120 - 2685015/720-840 CAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAA ..((((((.((((.(((.((..((((((((((((....(.((((((....)).....((((.(....))))))))).)..)))))))..)))))..)).)))))))..))))))...... ( -44.90) >sp_ag.0 1993425 120 - 2160267/720-840 CAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCGGCACUAAGCCCCGGAAAGGGCCUAACACCUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAA ..((((((.((((.(((.((..((((((((((((....(.(((((..(((((.....)))((.....))))))))).)..)))))))..)))))..)).)))))))..))))))...... ( -41.40) >consensus CAACCUUGCGGUCGUACUCCCCAGGCGGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAA ..((((((.((((.(((.((..((((((((((((....(.((((((....)).....((((......)))).)))).)..)))))))..)))))..)).)))))))..))))))...... (-41.88 = -40.75 + -1.12)

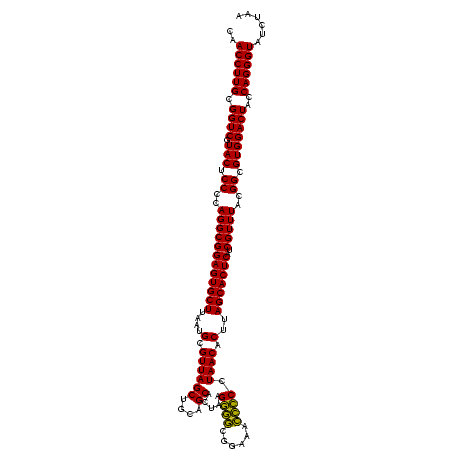
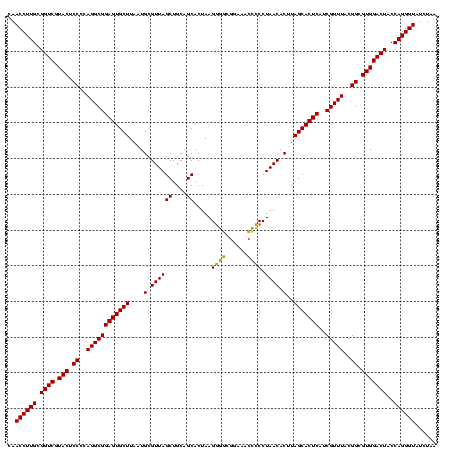
| Location | 1,980,435 – 1,980,555 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -50.37 |
| Consensus MFE | -51.88 |
| Energy contribution | -50.38 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.92 |
| Structure conservation index | 1.03 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 120 + 2809422/720-840 UUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUG .......(((.(((.........((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))).))).....(((((....))))) ( -49.80) >sy_sa.0 1555301 120 - 2516575/720-840 UUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUG .......(((.(((.........((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))).))).....(((((....))))) ( -49.80) >sy_ha.0 1580250 120 - 2685015/720-840 UUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUG .......(((.(((.........((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))).))).....(((((....))))) ( -49.80) >sp_ag.0 1993425 120 - 2160267/720-840 UUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGCCCUUUCCGGGGCUUAGUGCCGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUG .......(((.(((.........((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))).))).....(((((....))))) ( -52.10) >consensus UUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUG .......(((.(((.........((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))).))).....(((((....))))) (-51.88 = -50.38 + -1.50) # Strand winner: reverse (0.97)


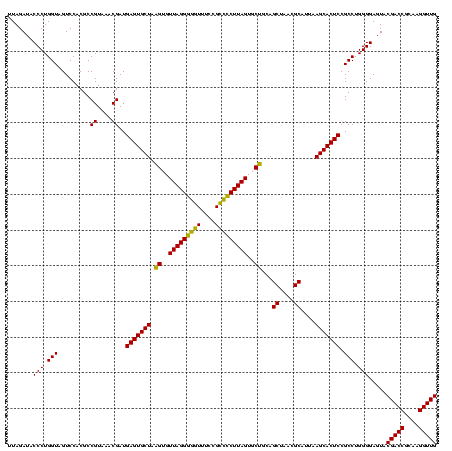
| Location | 1,980,435 – 1,980,555 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -43.05 |
| Consensus MFE | -43.19 |
| Energy contribution | -41.75 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.63 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860631 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 120 + 2809422/760-880 AACUGACGCUGAUGUGCGAAAGCGUGGGGAUCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAAC .(((.(((...(((.((....))(((.((((....((((.........))))...)))).))))))...)).).)))(((.((..(((((((((....)))))))))..))..))).... ( -41.90) >sy_sa.0 1555301 120 - 2516575/760-880 AACUGACGCUGAUGUGCGAAAGCGUGGGGAUCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAAC .(((.(((...(((.((....))(((.((((....((((.........))))...)))).))))))...)).).)))(((.((..(((((((((....)))))))))..))..))).... ( -41.90) >sy_ha.0 1580250 120 - 2685015/760-880 AACUGACGCUGAUGUGCGAAAGCGUGGGGAUCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAAC .(((.(((...(((.((....))(((.((((....((((.........))))...)))).))))))...)).).)))(((.((..(((((((((....)))))))))..))..))).... ( -41.90) >sp_ag.0 1993425 120 - 2160267/760-880 AACUGACGCUGAGGCUCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGCCCUUUCCGGGGCUUAGUGCCGCAGCUAAC .......((((..(((((...(((((((..((...((((.........))))))..)))))))((....)).)))))....((..(((((((((....)))))))))..)).)))).... ( -46.50) >consensus AACUGACGCUGAUGUGCGAAAGCGUGGGGAUCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAAC .......((((..((((....(((((((.......((((.........))))....)))))))((....))....))))..((..(((((((((....)))))))))..)).)))).... (-43.19 = -41.75 + -1.44) # Strand winner: reverse (0.99)

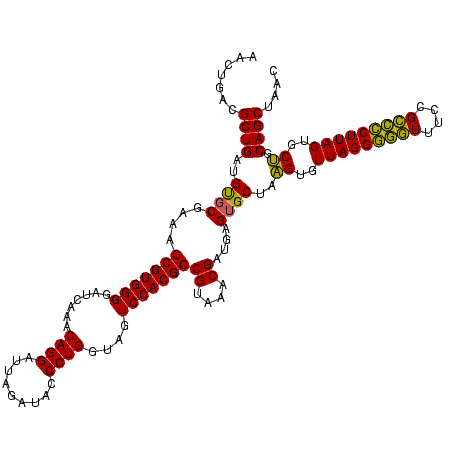
| Location | 1,980,435 – 1,980,555 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -38.88 |
| Consensus MFE | -36.89 |
| Energy contribution | -36.33 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.71 |
| SVM RNA-class probability | 0.973319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 120 + 2809422/840-960 AAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACGCUGAUGUGCGAAAGCGUGGGGAUCAAACAGGA ...(((...((((((((.(.(((........)))..).))))))))...))).(((.((....)).)))(((((...((...((.(((((..........))))).))...))..))))) ( -37.60) >sy_sa.0 1555301 120 - 2516575/840-960 AAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACGCUGAUGUGCGAAAGCGUGGGGAUCAAACAGGA ...(((...((((((((.(.(((........)))..).))))))))...))).(((.((....)).)))(((((...((...((.(((((..........))))).))...))..))))) ( -37.60) >sy_ha.0 1580250 120 - 2685015/840-960 AAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACGCUGAUGUGCGAAAGCGUGGGGAUCAAACAGGA ...(((...((((((((.(.(((........)))..).))))))))...))).(((.((....)).)))(((((...((...((.(((((..........))))).))...))..))))) ( -37.60) >sp_ag.0 1993425 120 - 2160267/840-960 AGAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAUAUAUGGAGGAACACCGGUGGCGAAAGCGGCUCUCUGGUCUGUAACUGACGCUGAGGCUCGAAAGCGUGGGGAGCAAACAGGA .(.(((...((((((((((.(((......)))....))))))))))...))))(((.((....)).))).((((...(((..((.(((((..........))))).))..)))..)))). ( -42.70) >consensus AAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACGCUGAUGUGCGAAAGCGUGGGGAUCAAACAGGA ...(((...((((((((.(.(((........)))..).))))))))...))).(((.((....)).))).((((...((...((.(((((..........))))).))...))..)))). (-36.89 = -36.33 + -0.56) # Strand winner: reverse (1.00)


| Location | 1,980,435 – 1,980,555 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.92 |
| Mean single sequence MFE | -37.97 |
| Consensus MFE | -34.40 |
| Energy contribution | -34.15 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992777 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 120 + 2809422/880-1000 GAGGGUCAUUGGAAACUGGAAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGU (((..((...(....)..))...)))....(((((..(((((.(((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).)))))))..))))) ( -36.80) >sy_sa.0 1555301 120 - 2516575/880-1000 GAGGGUCAUUGGAAACUGGGAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGU (((..((...(....)...))..)))....(((((..(((((.(((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).)))))))..))))) ( -39.00) >sy_ha.0 1580250 120 - 2685015/880-1000 GAGGGUCAUUGGAAACUGGAAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGU (((..((...(....)..))...)))....(((((..(((((.(((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).)))))))..))))) ( -36.80) >sp_ag.0 1993425 119 - 2160267/880-1000 GUACGCU-UUGGAAACUGGAGGACUUGAGUGCAGAAGGGGAGAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAUAUAUGGAGGAACACCGGUGGCGAAAGCGGCUCUCUGGUCUGU .....((-(((....).)))).........(((((.((((((.(((...((((((((((.(((......)))....))))))))))...)))).((.((....)).)))))))..))))) ( -39.30) >consensus GAGGGUCAUUGGAAACUGGAAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGCAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGU (((..((...(....)...))..)))....(((((...((((((((...((((((((.(.(((........)))..).))))))))...)))..((.((....)).)))))))..))))) (-34.40 = -34.15 + -0.25) # Strand winner: reverse (1.00)

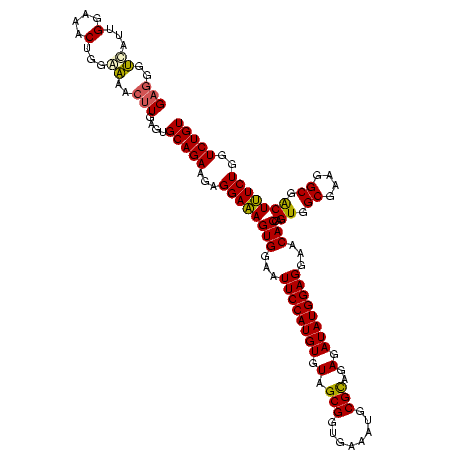
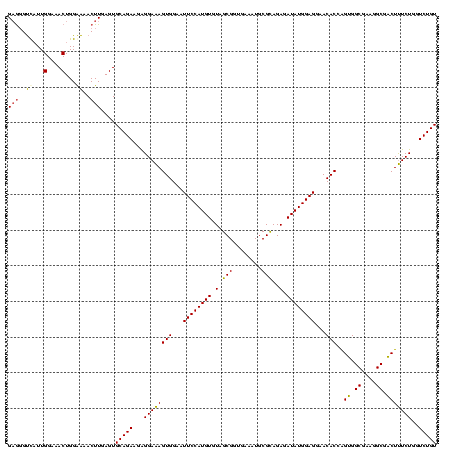
| Location | 1,980,435 – 1,980,554 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.59 |
| Mean single sequence MFE | -34.85 |
| Consensus MFE | -35.37 |
| Energy contribution | -32.56 |
| Covariance contribution | -2.81 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.68 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 119 + 2809422/1080-1200 UAAAACUCUGUUAUUAGGGAAGAACAUAUGUGUAAGU-AACUGUGCACAUCUUGACGGUACCUAAUCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAA ......((((..((((((......((.(((((((...-.....)))))))..))......))))))))))..(((((......(((((((((.((....)))))...))))))))))).. ( -34.00) >sy_sa.0 1555301 119 - 2516575/1080-1200 UAAAACUCUGUUAUUAGGGAAGAACAAACGUGUAAGU-AACUGUGCACGUCUUGACGGUACCUAAUCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAA ......((((..((((((......((((((((((...-.....))))))).)))......))))))))))..(((((......(((((((((.((....)))))...))))))))))).. ( -36.20) >sy_ha.0 1580250 119 - 2685015/1080-1200 UAAAACUCUGUUAUUAGGGAAGAACAUACGUGUAAGU-AACUAUGCACGUCUUGACGGUACCUAAUCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAA ......((((..((((((......((.(((((((...-.....)))))))..))......))))))))))..(((((......(((((((((.((....)))))...))))))))))).. ( -35.40) >sp_ag.0 1993425 120 - 2160267/1080-1200 UAAAGCUCUGUUGUUAGAGAAGAACGUUGGUAGGAGUGGAAAAUCUACCAAGUGACGGUAACUAACCAGAAAGGGACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUCCCGA .....(((((....)))))....((.((((((((..(....).))))))))))...(((.....))).....(((((......(((((((((.((....)))))...))))))))))).. ( -33.80) >consensus UAAAACUCUGUUAUUAGGGAAGAACAUACGUGUAAGU_AACUAUGCACGUCUUGACGGUACCUAAUCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAA ......((((..((((((.........(((((((.........)))))))..........))))))))))..(((((......(((((((((.((....)))))...))))))))))).. (-35.37 = -32.56 + -2.81) # Strand winner: reverse (0.97)

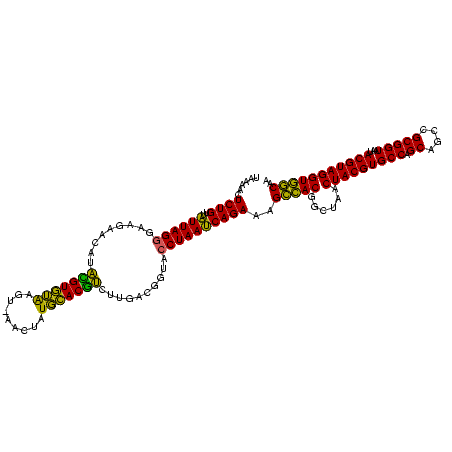
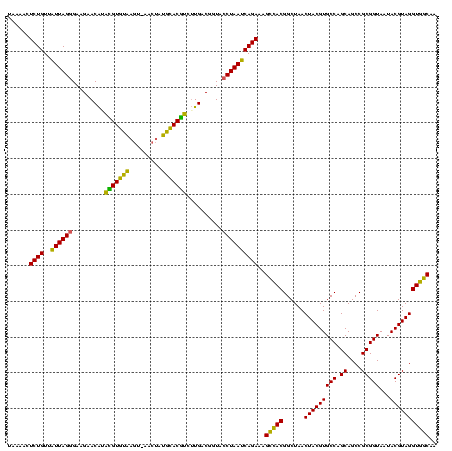
| Location | 1,980,435 – 1,980,554 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.79 |
| Mean single sequence MFE | -37.50 |
| Consensus MFE | -32.67 |
| Energy contribution | -32.18 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 119 + 2809422/1160-1280 AGGCAGCAGUAGGGAAUCUUCCGCAAUGGGCGAAAGCCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUCUUCGGAUCGUAAAACUCUGUUAUUAGGGAAGAACAUAUGUGUAAGU-AA ..(((.((....(...((((((.....((((....))))(((((((...((.(((....))).))..((((....))))......))))))).....)))))).)...)))))....-.. ( -39.20) >sy_sa.0 1555301 119 - 2516575/1160-1280 AGGCAGCAGUAGGGAAUCUUCCGCAAUGGGCGAAAGCCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGGUUUCGGCUCGUAAAACUCUGUUAUUAGGGAAGAACAAACGUGUAAGU-AA .....(((((..(...((((((.....((((....))))(((((((...((.(((....))).))..((((....))))......))))))).....)))))).)..)).)))....-.. ( -35.90) >sy_ha.0 1580250 119 - 2685015/1160-1280 AGGCAGCAGUAGGGAAUCUUCCGCAAUGGGCGAAAGCCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUCUUCGGAUCGUAAAACUCUGUUAUUAGGGAAGAACAUACGUGUAAGU-AA .....((((((.(...((((((.....((((....))))(((((((...((.(((....))).))..((((....))))......))))))).....)))))).).))).)))....-.. ( -40.50) >sp_ag.0 1993425 120 - 2160267/1160-1280 AGGCAGCAGUAGGGAAUCUUCGGCAAUGGACGGAAGUCUGACCGAGCAACGCCGCGUGAGUGAAGAAGGUUUUCGGAUCGUAAAGCUCUGUUGUUAGAGAAGAACGUUGGUAGGAGUGGA ...((((....(..((((((((((...((((....))))(......)...)))((....))...)))))))..).....((....(((((....)))))....))))))........... ( -34.40) >consensus AGGCAGCAGUAGGGAAUCUUCCGCAAUGGGCGAAAGCCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUCUUCGGAUCGUAAAACUCUGUUAUUAGGGAAGAACAUACGUGUAAGU_AA ................((((((.....((((....))))(((((((...(..(((....)))..)..((((....))))......))))))).....))))))................. (-32.67 = -32.18 + -0.50) # Strand winner: reverse (1.00)

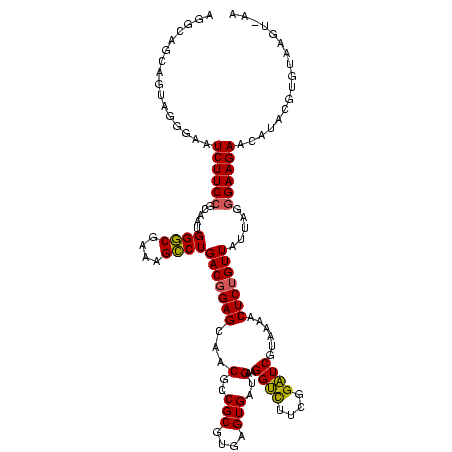
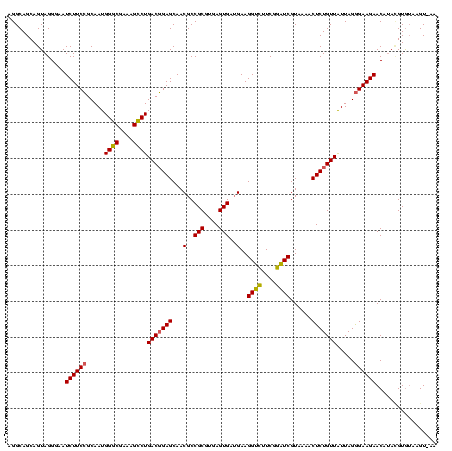
| Location | 1,980,435 – 1,980,555 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -45.73 |
| Consensus MFE | -42.94 |
| Energy contribution | -42.00 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 120 + 2809422/1240-1360 UACCAAGGCAACGAUGCAUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGGCGAAAGCCUG .......((.((..(((...((((((((....)))...)))))....(((((.(((.....)))))))).((((....)))))))...))..(((....)))))...((((....)))). ( -46.60) >sy_sa.0 1555301 120 - 2516575/1240-1360 UACCAAGGCAACGAUACGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGGCGAAAGCCUG .......((.....(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....))))......))).(((....)))))...((((....)))). ( -45.60) >sy_ha.0 1580250 120 - 2685015/1240-1360 UACCAAGGCGACGAUACGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGGCGAAAGCCUG .......((.....(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....))))......))).(((....)))))...((((....)))). ( -45.60) >sp_ag.0 1993425 120 - 2160267/1240-1360 CACCAAGGCGACGAUACAUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCGGCAAUGGACGGAAGUCUG ......(.(((.(((.(...((((((((....)))...))))).((.(((((.(((.....)))))))).((((....))))......))...).))).))).)...((((....)))). ( -45.10) >consensus UACCAAGGCAACGAUACAUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGAACUGAGACACGGUCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGGCGAAAGCCUG ..((..(....)..(((...((((((((....)))...)))))....(((((.(((.....)))))))).((((....))))......)))))..............((((....)))). (-42.94 = -42.00 + -0.94) # Strand winner: reverse (0.86)

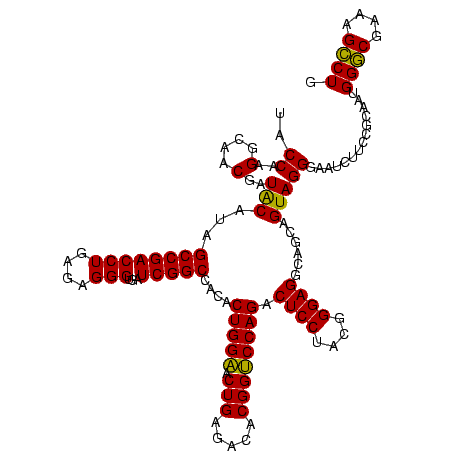
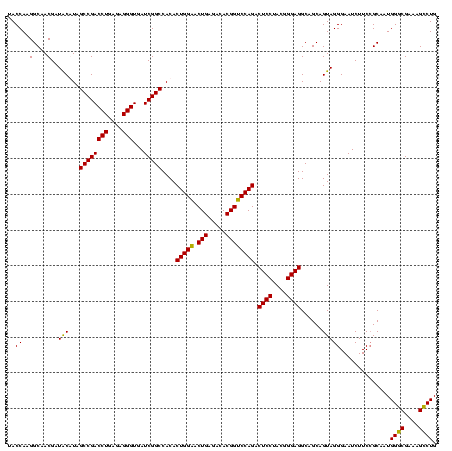
| Location | 1,980,435 – 1,980,555 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -43.60 |
| Consensus MFE | -42.20 |
| Energy contribution | -41.57 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 120 + 2809422/1280-1400 CCCGUAGGAGUCUGGACCGUGUCUCAGUUCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUAUGCAUCGUUGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUGCAGCGCGGAUCCA .((((.((((.((((((...))).))))))).((((((((((((.........))))))))))))...(((((.((((((((.......)))))))).........)))))))))..... ( -44.70) >sy_sa.0 1555301 120 - 2516575/1280-1400 CCCGUAGGAGUCUGGACCGUGUCUCAGUUCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGUAUCGUUGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUACGGCGCGGGUCCA (((((.((((.((((((...))).))))))).((((((((((((.........))))))))))((((....((.((((((((.......))))))))...))..)))).))))))).... ( -45.60) >sy_ha.0 1580250 120 - 2685015/1280-1400 CCCGUAGGAGUCUGGACCGUGUCUCAGUUCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGUAUCGUCGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUACGGCGCGGGUCCA (((((.((((.((((((...))).))))))).((((((((((((.........))))))))))((((....((.((((((((.......))))))))...))..)))).))))))).... ( -44.90) >sp_ag.0 1993425 120 - 2160267/1280-1400 CCCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUAUGUAUCGUCGCCUUGGUGAGCCUUUACCUCACCAACUAGCUAAUACAACGCAGGUCCA .((...((.(.(((((......))))).))).((((((((((((.........)))))))).(((((....((.((((((((.......))))))))...))..))))))))).)).... ( -39.20) >consensus CCCGUAGGAGUCUGGACCGUGUCUCAGUUCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGUAUCGUCGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUACAGCGCGGGUCCA (((((.((((.((((((...))).))))))).((((((((((((.........))))))))))((((....((.((((((((.......))))))))...))..)))).))))))).... (-42.20 = -41.57 + -0.63)

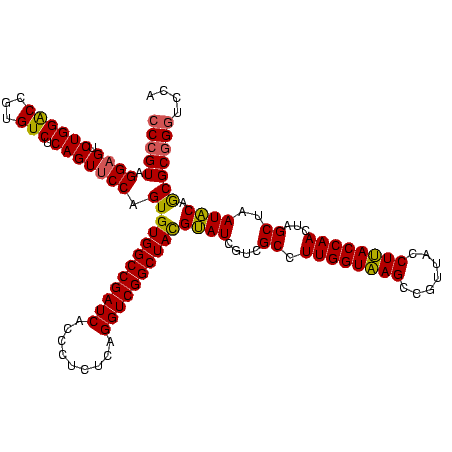
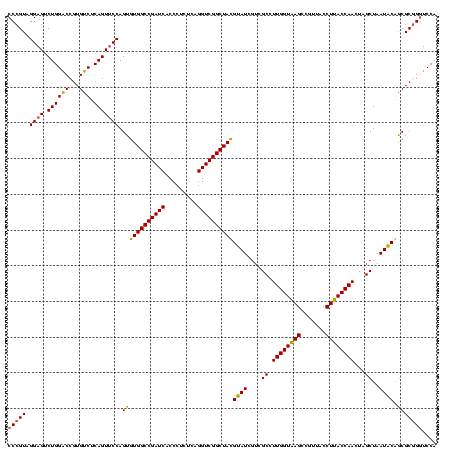
| Location | 1,980,435 – 1,980,555 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -22.46 |
| Energy contribution | -22.40 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 120 + 2809422/1360-1480 AGCCGUUACCUUACCAACUAGCUAAUGCAGCGCGGAUCCAUCUAUAAGUGACAGCAAGACCGUCUUUCACUUUUGAACCAUGCGGUUCAAAAUAUUAUCCGGUAUUAGCUCCGGUUUCCC (((((((........))).(((((((((....(((((..........((((.(((......).)).))))(((((((((....)))))))))....))))))))))))))..)))).... ( -31.60) >sy_sa.0 1555301 120 - 2516575/1360-1480 AGCCGUUACCUUACCAACUAGCUAAUACGGCGCGGGUCCAUCUAUAAGUGAUAGCAAAACCAUCUUUCACUUUAGAACCAUGCGGUUCCAAAUGUUAUCCGGUAUUAGCUCCGGUUUCCC (((((((........))).(((((((((.(.((((((...((((.((((((.((.........)).))))))))))))).)))(((..(....)..)))).)))))))))..)))).... ( -28.30) >sy_ha.0 1580250 120 - 2685015/1360-1480 AGCCGUUACCUUACCAACUAGCUAAUACGGCGCGGGUCCAUCUAUAAGUGAUAGCAAAACCAUCUUUCACUAUCGAACCAUGCGGUUCGAAAUAUUAUCCGGUAUUAGCUCCGGUUUCCC (((((((........))).(((((((((....(((((.........(((((.((.........)).))))).(((((((....)))))))......))))))))))))))..)))).... ( -31.20) >sp_ag.0 1993425 119 - 2160267/1360-1480 AGCCUUUACCUCACCAACUAGCUAAUACAACGCAGGUCCAUCUCACAGUGA-AGCAAUUGCUCCUUUUAAAUAACUAACAUGUGUUAAUUACUCUUAUGCGGUAUUAGCUAUCGUUUCCA ..................((((((((((..((((((......))..(((((-(((....))).........((((........)))).)))))....))))))))))))))......... ( -20.80) >consensus AGCCGUUACCUUACCAACUAGCUAAUACAGCGCGGGUCCAUCUAUAAGUGAUAGCAAAACCAUCUUUCACUUUAGAACCAUGCGGUUCAAAAUAUUAUCCGGUAUUAGCUCCGGUUUCCC ............(((....(((((((((....(((((.........(((((.((.........)).)))))((((((((....)))))))).....))))))))))))))..)))..... (-22.46 = -22.40 + -0.06)

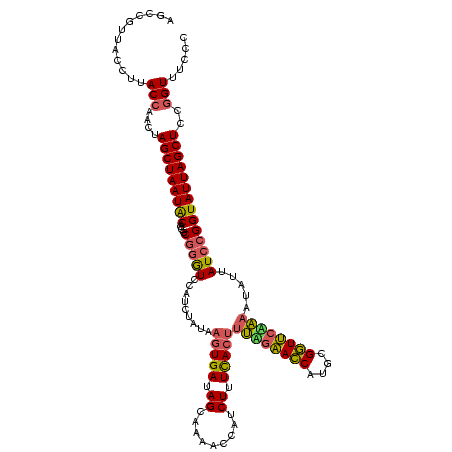
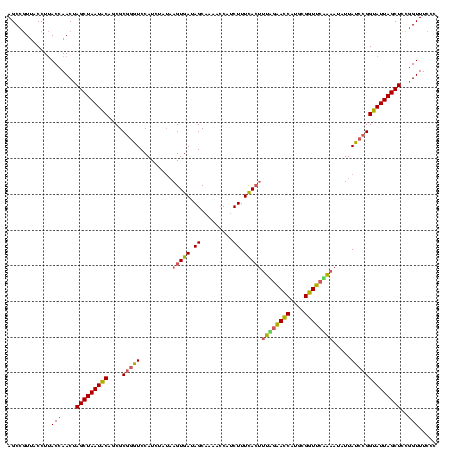
| Location | 1,980,435 – 1,980,555 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -36.20 |
| Consensus MFE | -31.67 |
| Energy contribution | -28.68 |
| Covariance contribution | -3.00 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.936755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 120 + 2809422/1360-1480 GGGAAACCGGAGCUAAUACCGGAUAAUAUUUUGAACCGCAUGGUUCAAAAGUGAAAGACGGUCUUGCUGUCACUUAUAGAUGGAUCCGCGCUGCAUUAGCUAGUUGGUAAGGUAACGGCU ((....))..((((..((((((((....(((((((((....)))))))))((((..(((((.....)))))..))))......))))..(((((....)).)))......))))..)))) ( -37.20) >sy_sa.0 1555301 120 - 2516575/1360-1480 GGGAAACCGGAGCUAAUACCGGAUAACAUUUGGAACCGCAUGGUUCUAAAGUGAAAGAUGGUUUUGCUAUCACUUAUAGAUGGACCCGCGCCGUAUUAGCUAGUUGGUAAGGUAACGGCU ((....))..(((((((((.((......(((((((((....)))))))))(((...(((((.....))))).((.......))...))).)))))))))))(((((.........))))) ( -36.70) >sy_ha.0 1580250 120 - 2685015/1360-1480 GGGAAACCGGAGCUAAUACCGGAUAAUAUUUCGAACCGCAUGGUUCGAUAGUGAAAGAUGGUUUUGCUAUCACUUAUAGAUGGACCCGCGCCGUAUUAGCUAGUUGGUAAGGUAACGGCU ((....))..(((((((((((((......))))...(((..((((((((((..(((.....)))..))))).(.....)..))))).)))..)))))))))(((((.........))))) ( -37.40) >sp_ag.0 1993425 119 - 2160267/1360-1480 UGGAAACGAUAGCUAAUACCGCAUAAGAGUAAUUAACACAUGUUAGUUAUUUAAAAGGAGCAAUUGCU-UCACUGUGAGAUGGACCUGCGUUGUAUUAGCUAGUUGGUGAGGUAAAGGCU .(....)..((((((((((((((...(((((((((((....)))))))))))....(((((....)))-))...............))))..)))))))))).................. ( -33.50) >consensus GGGAAACCGGAGCUAAUACCGGAUAAUAUUUAGAACCGCAUGGUUCUAAAGUGAAAGAUGGUUUUGCUAUCACUUAUAGAUGGACCCGCGCCGUAUUAGCUAGUUGGUAAGGUAACGGCU ((....))..((((..((((........(((((((((....)))))))))......(((((.....)))))............(((.((...((....))..)).)))..))))..)))) (-31.67 = -28.68 + -3.00) # Strand winner: reverse (1.00)

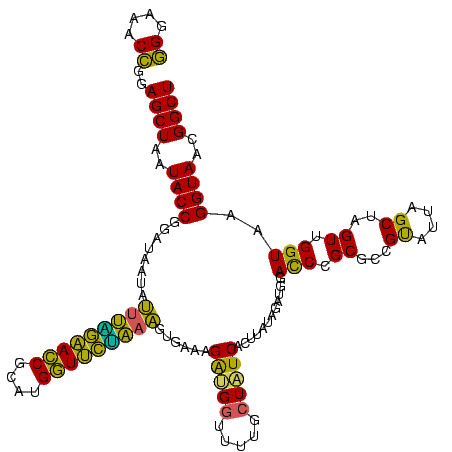
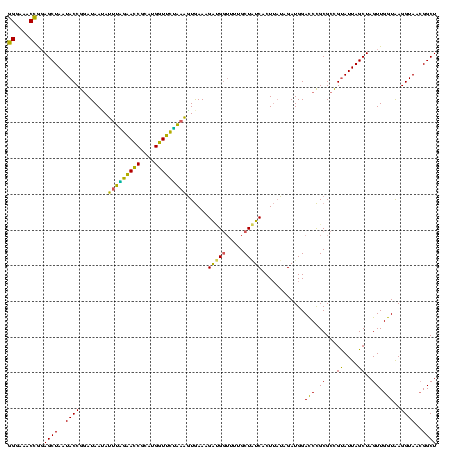
| Location | 1,980,435 – 1,980,555 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.86 |
| Mean single sequence MFE | -42.52 |
| Consensus MFE | -43.58 |
| Energy contribution | -38.58 |
| Covariance contribution | -5.00 |
| Combinations/Pair | 1.51 |
| Mean z-score | -4.77 |
| Structure conservation index | 1.02 |
| SVM decision value | 2.96 |
| SVM RNA-class probability | 0.997905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 120 + 2809422/1400-1520 UAACACGUGGAUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUUGAACCGCAUGGUUCAAAAGUGAAAGACGGUCUUGCUGUCACUUAUAGA ......((((.....))))(((((((.((((..((.((((((....)))))).))...))))......(((((((((....)))))))))......(((((.....))))).))))))). ( -42.50) >sy_sa.0 1555301 120 - 2516575/1400-1520 UAACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAACAUUUGGAACCGCAUGGUUCUAAAGUGAAAGAUGGUUUUGCUAUCACUUAUAGA ......(((((...)))))(((((((.((((..((.((((((....)))))).))...))))....(((((((((((....)))))).)))))...(((((.....))))).))))))). ( -43.20) >sy_ha.0 1580250 120 - 2685015/1400-1520 UGACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUCGAACCGCAUGGUUCGAUAGUGAAAGAUGGUUUUGCUAUCACUUAUAGA ......(((((...)))))(((((((.((((..((.((((((....)))))).))...))))..........(((((....)))))(((((..(((.....)))..))))).))))))). ( -43.40) >sp_ag.0 1993425 119 - 2160267/1400-1520 UAACGCGUAGGUAACCUGCCUCAUAGCGGGGGAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAGAGUAAUUAACACAUGUUAGUUAUUUAAAAGGAGCAAUUGCU-UCACUGUGAGA ......(((((...)))))((((((((((....((.((((((....)))))).))...))))....(((((((((((....)))))))))))....(((((....)))-))..)))))). ( -41.00) >consensus UAACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUAGAACCGCAUGGUUCUAAAGUGAAAGAUGGUUUUGCUAUCACUUAUAGA ......((((.....))))(((((((.((((..((.((((((....)))))).))...))))......(((((((((....)))))))))......(((((.....))))).))))))). (-43.58 = -38.58 + -5.00) # Strand winner: reverse (1.00)


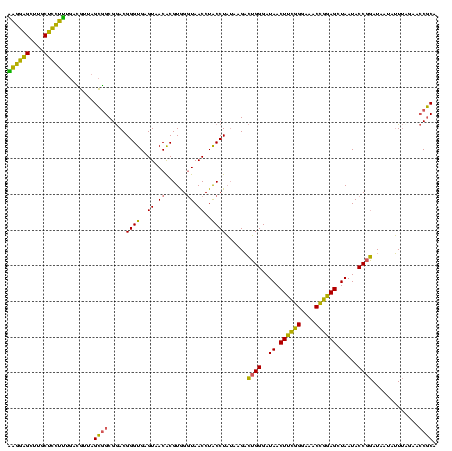
| Location | 1,980,435 – 1,980,555 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.31 |
| Mean single sequence MFE | -38.60 |
| Consensus MFE | -35.90 |
| Energy contribution | -32.90 |
| Covariance contribution | -3.00 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978552 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 120 + 2809422/1440-1560 GAGAAGCUUGCUUCUCUGAUGUUAGCGGCGGACGGGUGAGUAACACGUGGAUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUUGAACCGCA ((((((....))))))........((((.(((((.((.....)).)))((....))..)).......((((..((.((((((....)))))).))...)))).............)))). ( -36.90) >sy_sa.0 1555301 120 - 2516575/1440-1560 AAGGAGCUUGCUCCUUUGACGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAACAUUUGGAACCGCA ((((((....))))))...((....))((((.((((((........(((((((...)))))))....((((..((.((((((....)))))).))...))))....))))))...)))). ( -42.80) >sy_ha.0 1580250 120 - 2685015/1440-1560 AAGGAGCUUGCUCCUUUGACGUUAGCGGCGGACGGGUGAGUGACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUCGAACCGCA ((((((....))))))...((....))((((.((((((.....)))(((((((...)))))))....((((..((.((((((....)))))).))...))))........)))..)))). ( -40.00) >sp_ag.0 1993425 120 - 2160267/1440-1560 UUGGUGUUUACACUAGACUGAUGAGUUGCGAACGGGUGAGUAACGCGUAGGUAACCUGCCUCAUAGCGGGGGAUAACUAUUGGAAACGAUAGCUAAUACCGCAUAAGAGUAAUUAACACA ...((((((((.((.((((....))))((..(((.((.....)).))).((((.(((((......)))))......((((((....))))))....))))))...)).)))...))))). ( -34.70) >consensus AAGGAGCUUGCUCCUUUGACGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUACCUAUAAGACUGGGAUAACUUCGGGAAACCGGAGCUAAUACCGGAUAAUAUUUAGAACCGCA ((((((....))))))........((((.....((((..((.((......)).))..))))......((((..((.((((((....)))))).))...)))).............)))). (-35.90 = -32.90 + -3.00) # Strand winner: reverse (1.00)

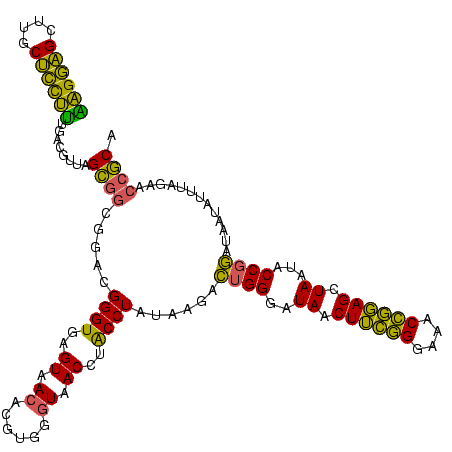
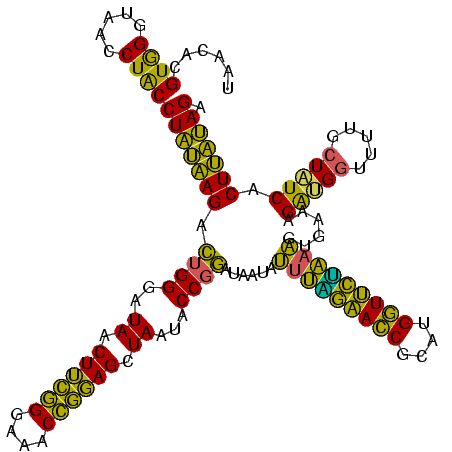
| Location | 1,980,435 – 1,980,551 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.07 |
| Mean single sequence MFE | -32.55 |
| Consensus MFE | -20.61 |
| Energy contribution | -22.93 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.733463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 116 + 2809422/1560-1680 -AACAUUAUUUAGUA-UUUAUGAGCUAAUCAAACAUCAUAAUU--UUUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGAACGGAC -............((-(((.((((((((((((((.(((((...--..)))))...)))))))..))))))))))))..(((((((.(((((.......)))))..))).))))....... ( -31.00) >sy_sa.0 1555301 120 - 2516575/1560-1680 UAACACAAAUUAGUAUUUUAUGAGCUAAUCAAACAUCAUAAUUCAUUUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGAACAGAU .............((((...((((((((((((((.((((((.....))))))...)))))))..))))))).))))..(((((((.(((((.......)))))..))).))))....... ( -32.40) >sy_ha.0 1580250 116 - 2685015/1560-1680 -AAAACUAUUUAGUA--UUAUGAGCUAAUCAAACAUCAUAAAUU-UUUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGAACAGAC -............((--((.((((((((((((((.((((((...-.))))))...)))))))..))))))).))))..(((((((.(((((.......)))))..))).))))....... ( -32.90) >sp_ag.0 1993425 116 - 2160267/1560-1680 AUAAAUCUGUCAGCGAGACA-GAAAAGAGCAAAGCUCA-AACU--UUUAAUGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUAGAACGCUGAGGU .....((((((.....))))-))...((((...(.(((-((((--(((...)))))))))).)...)))).........(((..((((((......(((......)))..)))))).))) ( -33.90) >consensus _AAAACUAUUUAGUA_UUUAUGAGCUAAUCAAACAUCAUAAUU__UUUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGAACAGAC .....................(((((((((((((.((((((.....))))))...)))))))..))))))........(((((((.(((((.......)))))..))).))))....... (-20.61 = -22.93 + 2.31) # Strand winner: reverse (0.99)

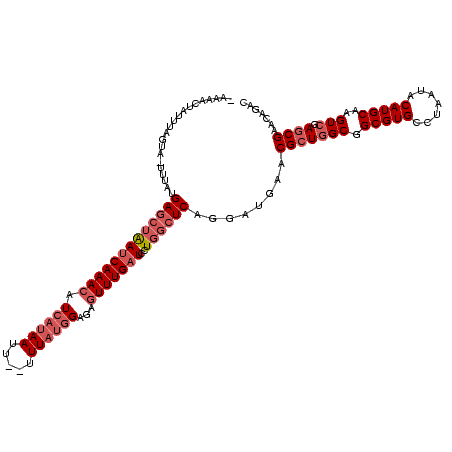

| Location | 1,980,435 – 1,980,531 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.18 |
| Mean single sequence MFE | -24.80 |
| Consensus MFE | -10.39 |
| Energy contribution | -11.58 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.42 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986290 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 96 + 2809422/1600-1720 CGUUCAUCCUGAGCCAGGAUCAAACUCUCCAUAAA--AAUUAUGAUGUUUGAUUAGCUCAUAAA-UACUAAAUAAUGUU------------UGU----AACUUAUAGUU--ACGUU---U .........(((((...((((((((....((((..--...))))..)))))))).)))))....-..............------------.((----(((.....)))--))...---. ( -20.10) >sy_sa.0 1555301 120 - 2516575/1600-1720 CGUUCAUCCUGAGCCAGGAUCAAACUCUCCAUAAAUGAAUUAUGAUGUUUGAUUAGCUCAUAAAAUACUAAUUUGUGUUAUUUCUAACACUUGUUUUGAACCAACAAUUUAAUGUUGUGU .(((((...(((((...((((((((....(((((.....)))))..)))))))).))))).........(((..((((((....))))))..))).)))))(((((......)))))... ( -29.30) >sy_ha.0 1580250 97 - 2685015/1600-1720 CGUUCAUCCUGAGCCAGGAUCAAACUCUCCAUAAA-AAUUUAUGAUGUUUGAUUAGCUCAUAA--UACUAAAUAGUUUU------------UGU----AACAUUUAAGUUACUGUU---- .........(((((...((((((((....(((((.-...)))))..)))))))).)))))...--..............------------.((----(((......)))))....---- ( -20.50) >sp_ag.0 1993425 110 - 2160267/1600-1720 CGUUCGUCCUGAGCCAGGAUCAAACUCUCAUUAAA--AGUU-UGAGCUUUGCUCUUUUC-UGUCUCGCUGACAGAUUUAUUGUUU----UUUGUCAUUGACGGAUU-UACAAUGUAA-AU ..........((((.(((.(((((((.........--))))-))).))).))))...((-((((.....))))))..((((((..----((((((...))))))..-.))))))...-.. ( -29.30) >consensus CGUUCAUCCUGAGCCAGGAUCAAACUCUCCAUAAA__AAUUAUGAUGUUUGAUUAGCUCAUAAA_UACUAAAUAAUGUU____________UGU____AACAUAUAAUUUAAUGUU___U .((......(((((...((((((((....(((((.....)))))..)))))))).)))))......)).................................................... (-10.39 = -11.58 + 1.19)

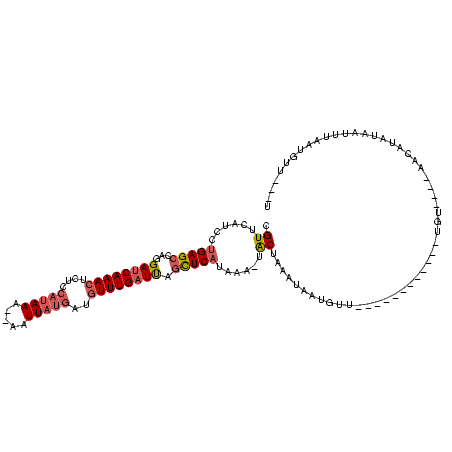
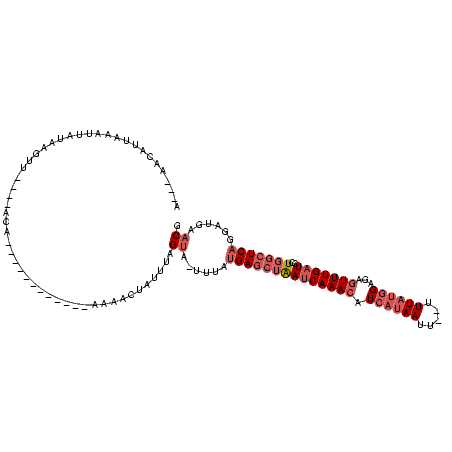
| Location | 1,980,435 – 1,980,531 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.18 |
| Mean single sequence MFE | -28.02 |
| Consensus MFE | -10.06 |
| Energy contribution | -12.38 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.82 |
| Structure conservation index | 0.36 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1980435 96 + 2809422/1600-1720 A---AACGU--AACUAUAAGUU----ACA------------AACAUUAUUUAGUA-UUUAUGAGCUAAUCAAACAUCAUAAUU--UUUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAACG .---...((--(((.....)))----)).------------............((-(((.((((((((((((((.(((((...--..)))))...)))))))..)))))))))))).... ( -22.10) >sy_sa.0 1555301 120 - 2516575/1600-1720 ACACAACAUUAAAUUGUUGGUUCAAAACAAGUGUUAGAAAUAACACAAAUUAGUAUUUUAUGAGCUAAUCAAACAUCAUAAUUCAUUUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAACG ...(((((......)))))(((((......((((((....))))))..............((((((((((((((.((((((.....))))))...)))))))..)))))))...))))). ( -36.70) >sy_ha.0 1580250 97 - 2685015/1600-1720 ----AACAGUAACUUAAAUGUU----ACA------------AAAACUAUUUAGUA--UUAUGAGCUAAUCAAACAUCAUAAAUU-UUUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAACG ----....(((((......)))----)).------------............((--((.((((((((((((((.((((((...-.))))))...)))))))..))))))).)))).... ( -24.40) >sp_ag.0 1993425 110 - 2160267/1600-1720 AU-UUACAUUGUA-AAUCCGUCAAUGACAAA----AAACAAUAAAUCUGUCAGCGAGACA-GAAAAGAGCAAAGCUCA-AACU--UUUAAUGAGAGUUUGAUCCUGGCUCAGGACGAACG ((-((((...)))-))).((((.........----..........((((((.....))))-))...((((...(.(((-((((--(((...)))))))))).)...))))..)))).... ( -28.90) >consensus A___AACAUUAAAUUAUAAGUU____ACA____________AAAACUAUUUAGUA_UUUAUGAGCUAAUCAAACAUCAUAAUU__UUUAUGGAGAGUUUGAUCCUGGCUCAGGAUGAACG ....................................................((......((((((((((((((.((((((.....))))))...)))))))..)))))))......)). (-10.06 = -12.38 + 2.31) # Strand winner: reverse (0.89)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:56:16 2006