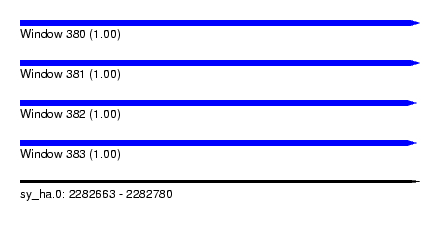
| Sequence ID | sy_ha.0 |
|---|---|
| Location | 2,282,663 – 2,282,780 |
| Length | 117 |
| Max. P | 0.999984 |

| Location | 2,282,663 – 2,282,780 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.55 |
| Mean single sequence MFE | -30.57 |
| Consensus MFE | -21.17 |
| Energy contribution | -20.85 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.31 |
| Mean z-score | -5.31 |
| Structure conservation index | 0.69 |
| SVM decision value | 4.62 |
| SVM RNA-class probability | 0.999930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
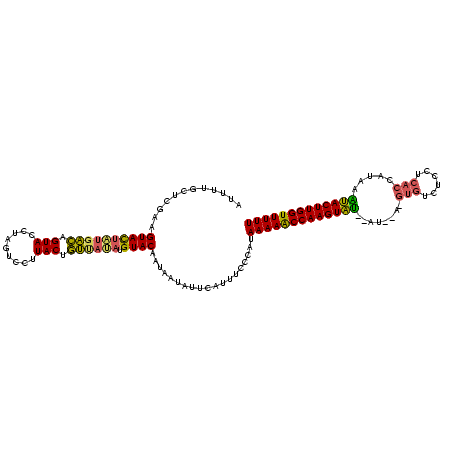
>sy_ha.0 2282663 117 + 2685015/0-120 AUUUUGCUCGAAGUACUGUGACAGUACCUAGUCCUUACCGUUACAAGUACAUUAAUAUUCAUUUCCCAUAAAAACCAAGUAU--AUAGA-GUGUCUCCUCACCCUACAUACUUGGUUUUU ............((((((((((.(((.........))).))))).)))))...................(((((((((((((--.(((.-(((......))).))).))))))))))))) ( -31.90) >sy_au.0 2081207 105 - 2809422/0-120 AUUUUGCUCGAAGUACUAUGACUGUACCUAGUCCUUACUGUCAUAUGUACAAUAAUAUUCAUUUCCCAUAAAAGCCAAGUGU--AU-------------UACCAUGUGCACUUGGUUUUU ............((((((((((.(((.........))).)))))).))))...................(((((((((((((--((-------------......))))))))))))))) ( -30.40) >sy_sa.0 2141536 117 + 2516575/0-120 UUUUUGCUCGA-GUAUAUUUUUAGUACCUAGUCCUUACUAAU-UAUGUACAAUUA-AUUCAUUUCCCGUAAAAACCAAGUAAUGAUGAAUGUGUCUCCUCACUUCAAUUACUUGGUUUUU ...........-((((((..((((((.........)))))).-.)))))).....-.............((((((((((((((..((((.(((......))))))))))))))))))))) ( -29.40) >consensus AUUUUGCUCGAAGUACUAUGACAGUACCUAGUCCUUACUGUUAUAUGUACAAUAAUAUUCAUUUCCCAUAAAAACCAAGUAU__AU__A_GUGUCUCCUCACCAUAAAUACUUGGUUUUU ............((((((((((.(((.........))).)))))).))))...................(((((((((((((........(((......))).....))))))))))))) (-21.17 = -20.85 + -0.32)

| Location | 2,282,663 – 2,282,780 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.55 |
| Mean single sequence MFE | -28.73 |
| Consensus MFE | -16.61 |
| Energy contribution | -19.07 |
| Covariance contribution | 2.46 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.58 |
| SVM decision value | 4.67 |
| SVM RNA-class probability | 0.999937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
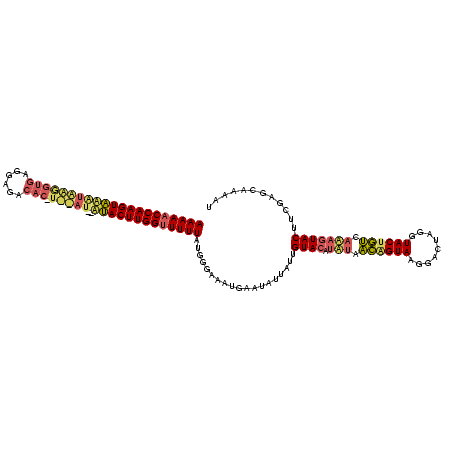
>sy_ha.0 2282663 117 + 2685015/0-120 AAAAACCAAGUAUGUAGGGUGAGGAGACAC-UCUAU--AUACUUGGUUUUUAUGGGAAAUGAAUAUUAAUGUACUUGUAACGGUAAGGACUAGGUACUGUCACAGUACUUCGAGCAAAAU (((((((((((((((((((((......)))-)))))--)))))))))))))...................(((((.((.((((((.........)))))).)))))))............ ( -39.30) >sy_au.0 2081207 105 - 2809422/0-120 AAAAACCAAGUGCACAUGGUA-------------AU--ACACUUGGCUUUUAUGGGAAAUGAAUAUUAUUGUACAUAUGACAGUAAGGACUAGGUACAGUCAUAGUACUUCGAGCAAAAU ((((.(((((((.........-------------..--.))))))).))))...................((((.((((((.(((.........))).))))))))))............ ( -19.50) >sy_sa.0 2141536 117 + 2516575/0-120 AAAAACCAAGUAAUUGAAGUGAGGAGACACAUUCAUCAUUACUUGGUUUUUACGGGAAAUGAAU-UAAUUGUACAUA-AUUAGUAAGGACUAGGUACUAAAAAUAUAC-UCGAGCAAAAA ((((((((((((((((((.((.(....).))))))..)))))))))))))).((((.(((....-..)))((((...-.(((((....)))))))))..........)-)))........ ( -27.40) >consensus AAAAACCAAGUAAAUAAGGUGAGGAGACAC_U__AU__AUACUUGGUUUUUAUGGGAAAUGAAUAUUAUUGUACAUAUAACAGUAAGGACUAGGUACUGUCAAAGUACUUCGAGCAAAAU (((((((((((((((((((((......))).)))))..)))))))))))))...................((((.(((.((((((.........)))))).)))))))............ (-16.61 = -19.07 + 2.46) # Strand winner: forward (0.97)

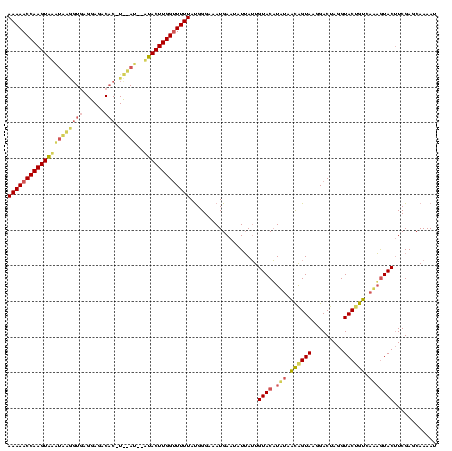
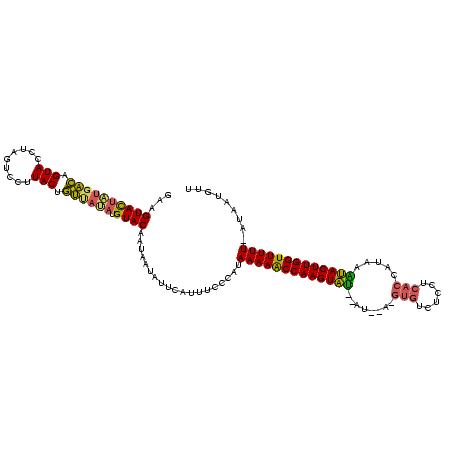
| Location | 2,282,663 – 2,282,779 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.51 |
| Mean single sequence MFE | -32.63 |
| Consensus MFE | -21.21 |
| Energy contribution | -20.89 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.31 |
| Mean z-score | -5.74 |
| Structure conservation index | 0.65 |
| SVM decision value | 3.97 |
| SVM RNA-class probability | 0.999733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2282663 116 + 2685015/9-129 GAAGUACUGUGACAGUACCUAGUCCUUACCGUUACAAGUACAUUAAUAUUCAUUUCCCAUAAAAACCAAGUAU--AUAGA-GUGUCUCCUCACCCUACAUACUUGGUUUUU-AUCAUGUU ...((((((((((.(((.........))).))))).))))).................(((((((((((((((--.(((.-(((......))).))).)))))))))))))-))...... ( -34.50) >sy_au.0 2081207 104 - 2809422/9-129 GAAGUACUAUGACUGUACCUAGUCCUUACUGUCAUAUGUACAAUAAUAUUCAUUUCCCAUAAAAGCCAAGUGU--AU-------------UACCAUGUGCACUUGGUUUUU-AUAAUGCU ...((((((((((.(((.........))).)))))).)))).................(((((((((((((((--((-------------......)))))))))))))))-))...... ( -33.60) >sy_sa.0 2141536 117 + 2516575/9-129 GA-GUAUAUUUUUAGUACCUAGUCCUUACUAAU-UAUGUACAAUUA-AUUCAUUUCCCGUAAAAACCAAGUAAUGAUGAAUGUGUCUCCUCACUUCAAUUACUUGGUUUUUUAUUGUGUU ..-((((((..((((((.........)))))).-.)))))).....-...(((.......((((((((((((((..((((.(((......)))))))))))))))))))))....))).. ( -29.80) >consensus GAAGUACUAUGACAGUACCUAGUCCUUACUGUUAUAUGUACAAUAAUAUUCAUUUCCCAUAAAAACCAAGUAU__AU__A_GUGUCUCCUCACCAUAAAUACUUGGUUUUU_AUAAUGUU ...((((((((((.(((.........))).)))))).))))...................(((((((((((((........(((......))).....)))))))))))))......... (-21.21 = -20.89 + -0.32)

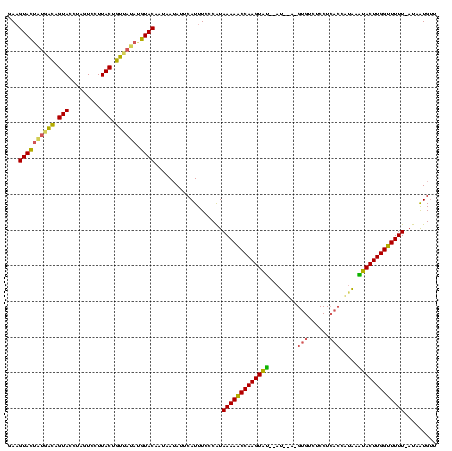
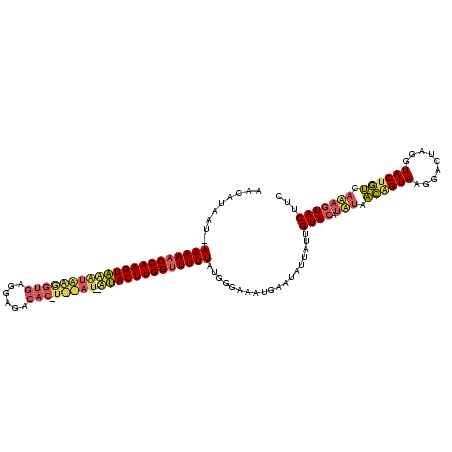
| Location | 2,282,663 – 2,282,779 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.51 |
| Mean single sequence MFE | -30.30 |
| Consensus MFE | -16.71 |
| Energy contribution | -19.17 |
| Covariance contribution | 2.46 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.73 |
| Structure conservation index | 0.55 |
| SVM decision value | 5.34 |
| SVM RNA-class probability | 0.999984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 2282663 116 + 2685015/9-129 AACAUGAU-AAAAACCAAGUAUGUAGGGUGAGGAGACAC-UCUAU--AUACUUGGUUUUUAUGGGAAAUGAAUAUUAAUGUACUUGUAACGGUAAGGACUAGGUACUGUCACAGUACUUC ....(.((-(((((((((((((((((((((......)))-)))))--))))))))))))))).)...............(((((.((.((((((.........)))))).)))))))... ( -42.50) >sy_au.0 2081207 104 - 2809422/9-129 AGCAUUAU-AAAAACCAAGUGCACAUGGUA-------------AU--ACACUUGGCUUUUAUGGGAAAUGAAUAUUAUUGUACAUAUGACAGUAAGGACUAGGUACAGUCAUAGUACUUC ....((((-((((.(((((((.........-------------..--.))))))).))))))))...............((((.((((((.(((.........))).))))))))))... ( -23.80) >sy_sa.0 2141536 117 + 2516575/9-129 AACACAAUAAAAAACCAAGUAAUUGAAGUGAGGAGACACAUUCAUCAUUACUUGGUUUUUACGGGAAAUGAAU-UAAUUGUACAUA-AUUAGUAAGGACUAGGUACUAAAAAUAUAC-UC .........((((((((((((((((((.((.(....).))))))..)))))))))))))).............-.....((((...-.(((((....)))))))))...........-.. ( -24.60) >consensus AACAUAAU_AAAAACCAAGUAAAUAAGGUGAGGAGACAC_U__AU__AUACUUGGUUUUUAUGGGAAAUGAAUAUUAUUGUACAUAUAACAGUAAGGACUAGGUACUGUCAAAGUACUUC .........(((((((((((((((((((((......))).)))))..)))))))))))))...................((((.(((.((((((.........)))))).)))))))... (-16.71 = -19.17 + 2.46) # Strand winner: forward (0.98)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:55:53 2006