| Sequence ID | sy_sa.0 |
|---|---|
| Location | 1,948,487 – 1,948,602 |
| Length | 115 |
| Max. P | 0.999993 |

| Location | 1,948,487 – 1,948,602 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.94 |
| Mean single sequence MFE | -24.21 |
| Consensus MFE | -15.09 |
| Energy contribution | -13.93 |
| Covariance contribution | -1.15 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.62 |
| SVM decision value | 3.82 |
| SVM RNA-class probability | 0.999638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
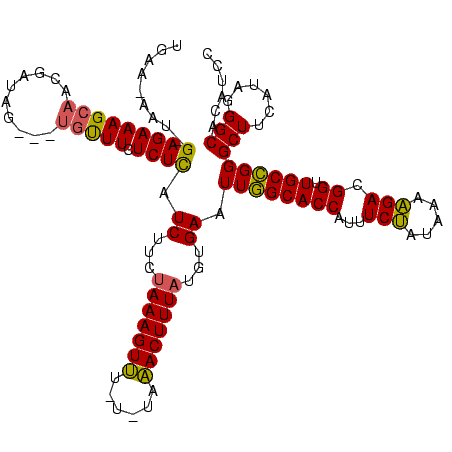
>sy_sa.0 1948487 115 + 2516575/0-120 AAAAAAUGCUUUCCCUAUGUUCAUUGAAGAAUAGAGAAAGCAACGUU-----UGCUUCUCUCAUCUUCUAAAGUUUCUAUGAACUUUAUGUGAAUUGGCACCAUUUCUGUUACAGACGGU .............((...((((((.(((((..((((((.(((.....-----)))))))))..)))))((((((((....)))))))).)))))).)).(((...((((...)))).))) ( -26.70) >sy_au.0 1908658 116 - 2809422/0-120 AAAAAAUGCUUUCUCAAUAUCGAUAGAAAAAUUGAGAAAGCAAUAGUAGUAUUGUUUCUCUCAUCUUCAAAAGUUA----AAACUUUAUGUGAAUUGGCACCAUUUCUAUAUAAGACGGU ......((((((((((((.((....))...))))))))))))........(((((((............(((((..----..)))))(((((((.((....)).))).)))).))))))) ( -23.30) >sy_ha.0 2209611 106 - 2685015/0-120 AAAAAAUACUUUC----UAUUAUUUAA------GAGAAAGUAUCGAUAG---ACUUUCUCUUAUCUCUUAAAGUAU-UGUCUACUUUAUGUGAAUUAGCACCAUUUCGUUAAACGACGGU ....((((((((.----.......(((------((((((((........---)))))))))))......)))))))-)((((((.....)))..(((((........)))))..)))... ( -22.64) >consensus AAAAAAUGCUUUC_C_AUAUUAAUUGAA_AAU_GAGAAAGCAACGAUAG___UGUUUCUCUCAUCUUCUAAAGUUU_U_UAAACUUUAUGUGAAUUGGCACCAUUUCUAUAAAAGACGGU ......(((((((.((((.((....))...)))).)))))))...........(((....((((....(((((((......))))))).))))...)))(((...(((.....))).))) (-15.09 = -13.93 + -1.15)

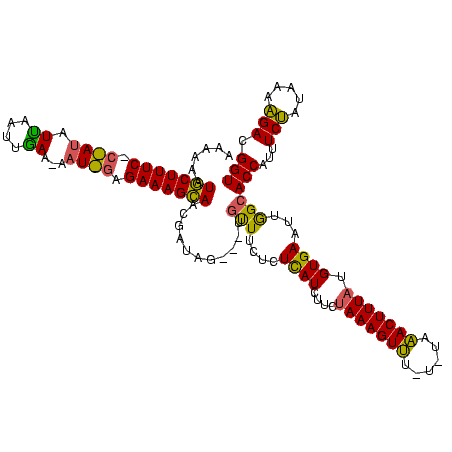
| Location | 1,948,487 – 1,948,602 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.94 |
| Mean single sequence MFE | -25.43 |
| Consensus MFE | -18.46 |
| Energy contribution | -18.30 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.73 |
| SVM decision value | 5.77 |
| SVM RNA-class probability | 0.999993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 1948487 115 + 2516575/0-120 ACCGUCUGUAACAGAAAUGGUGCCAAUUCACAUAAAGUUCAUAGAAACUUUAGAAGAUGAGAGAAGCA-----AACGUUGCUUUCUCUAUUCUUCAAUGAACAUAGGGAAAGCAUUUUUU ..(((((......(((.((....)).)))...(((((((......)))))))..))))).........-----.....((((((((((((((......))..))))))))))))...... ( -26.60) >sy_au.0 1908658 116 - 2809422/0-120 ACCGUCUUAUAUAGAAAUGGUGCCAAUUCACAUAAAGUUU----UAACUUUUGAAGAUGAGAGAAACAAUACUACUAUUGCUUUCUCAAUUUUUCUAUCGAUAUUGAGAAAGCAUUUUUU ..((((((.....(((.((....)).))).((.(((((..----..)))))))))))))...................((((((((((((...((....)).))))))))))))...... ( -26.30) >sy_ha.0 2209611 106 - 2685015/0-120 ACCGUCGUUUAACGAAAUGGUGCUAAUUCACAUAAAGUAGACA-AUACUUUAAGAGAUAAGAGAAAGU---CUAUCGAUACUUUCUC------UUAAAUAAUA----GAAAGUAUUUUUU (((.(((.....)))...)))(((..(((...(((((((....-.))))))).....(((((((((((---........))))))))------))).......----))))))....... ( -23.40) >consensus ACCGUCUUUUAAAGAAAUGGUGCCAAUUCACAUAAAGUUCA_A_AAACUUUAGAAGAUGAGAGAAACA___CUAACGUUGCUUUCUC_AUU_UUCAAUCAAUAU_G_GAAAGCAUUUUUU (((.(((.....)))...)))......((...(((((((......)))))))...)).....................((((((((((((............))))))))))))...... (-18.46 = -18.30 + -0.16) # Strand winner: reverse (0.94)

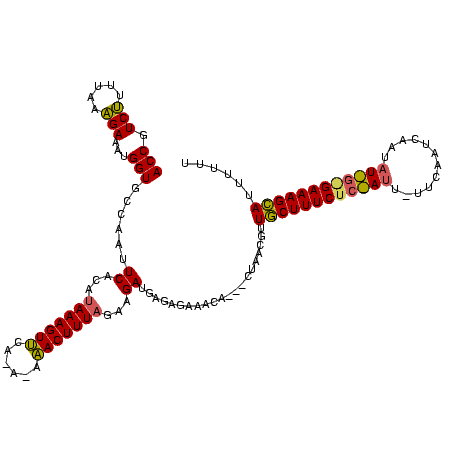
| Location | 1,948,487 – 1,948,602 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.88 |
| Mean single sequence MFE | -29.50 |
| Consensus MFE | -20.50 |
| Energy contribution | -19.73 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.993971 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 1948487 115 + 2516575/24-144 UGAAGAAUAGAGAAAGCAACGUU-----UGCUUCUCUCAUCUUCUAAAGUUUCUAUGAACUUUAUGUGAAUUGGCACCAUUUCUGUUACAGACGGUUGCCGGGCUUCAAAGGGCACAUCC .(((((..((((((.(((.....-----)))))))))..)))))((((((((....))))))))(((...((((((((...((((...)))).)).))))))(((......))))))... ( -32.00) >sy_au.0 1908658 116 - 2809422/24-144 AGAAAAAUUGAGAAAGCAAUAGUAGUAUUGUUUCUCUCAUCUUCAAAAGUUA----AAACUUUAUGUGAAUUGGCACCAUUUCUAUAUAAGACGGUUGCCGGGCUUCGUAGGGCACAUCC .(((....(((((((((((((....)))))))).)))))..)))........----...(((((((....((((((((...(((.....))).)).))))))....)))))))....... ( -27.00) >sy_ha.0 2209611 110 - 2685015/24-144 UAA------GAGAAAGUAUCGAUAG---ACUUUCUCUUAUCUCUUAAAGUAU-UGUCUACUUUAUGUGAAUUAGCACCAUUUCGUUAAACGACGGUUGCUGGGCUUCUUAGGGCACAUCC (((------((((((((........---)))))))))))((.(.(((((((.-....))))))).).)).((((((((...(((.....))).)).))))))(((......)))...... ( -29.50) >consensus UGAA_AAU_GAGAAAGCAACGAUAG___UGUUUCUCUCAUCUUCUAAAGUUU_U_UAAACUUUAUGUGAAUUGGCACCAUUUCUAUAAAAGACGGUUGCCGGGCUUCAUAGGGCACAUCC .........(((((((((..........))))).)))).((...(((((((......)))))))...)).((((((((...(((.....))).)).))))))(((......)))...... (-20.50 = -19.73 + -0.77)

| Location | 1,948,487 – 1,948,602 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.88 |
| Mean single sequence MFE | -27.63 |
| Consensus MFE | -21.25 |
| Energy contribution | -20.26 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.77 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 1948487 115 + 2516575/24-144 GGAUGUGCCCUUUGAAGCCCGGCAACCGUCUGUAACAGAAAUGGUGCCAAUUCACAUAAAGUUCAUAGAAACUUUAGAAGAUGAGAGAAGCA-----AACGUUGCUUUCUCUAUUCUUCA ((......))..((((....((((.((.((((...))))...))))))..))))..(((((((......)))))))(((((..(((((((((-----.....))).))))))..))))). ( -32.10) >sy_au.0 1908658 116 - 2809422/24-144 GGAUGUGCCCUACGAAGCCCGGCAACCGUCUUAUAUAGAAAUGGUGCCAAUUCACAUAAAGUUU----UAACUUUUGAAGAUGAGAGAAACAAUACUACUAUUGCUUUCUCAAUUUUUCU ..(((((......(.....)((((.((.(((.....)))...))))))....)))))(((((..----..))))).(((((...(((((((((((....))))).))))))...))))). ( -23.90) >sy_ha.0 2209611 110 - 2685015/24-144 GGAUGUGCCCUAAGAAGCCCAGCAACCGUCGUUUAACGAAAUGGUGCUAAUUCACAUAAAGUAGACA-AUACUUUAAGAGAUAAGAGAAAGU---CUAUCGAUACUUUCUC------UUA ((......))...(((....((((.((.(((.....)))...))))))..)))...(((((((....-.))))))).....(((((((((((---........))))))))------))) ( -26.90) >consensus GGAUGUGCCCUAAGAAGCCCGGCAACCGUCUUUUAAAGAAAUGGUGCCAAUUCACAUAAAGUUCA_A_AAACUUUAGAAGAUGAGAGAAACA___CUAACGUUGCUUUCUC_AUU_UUCA (((((.((........)).)((((.((.(((.....)))...)))))).))))...(((((((......)))))))(((((...((((((...............))))))...))))). (-21.25 = -20.26 + -0.99) # Strand winner: forward (0.69)


Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:55:39 2006