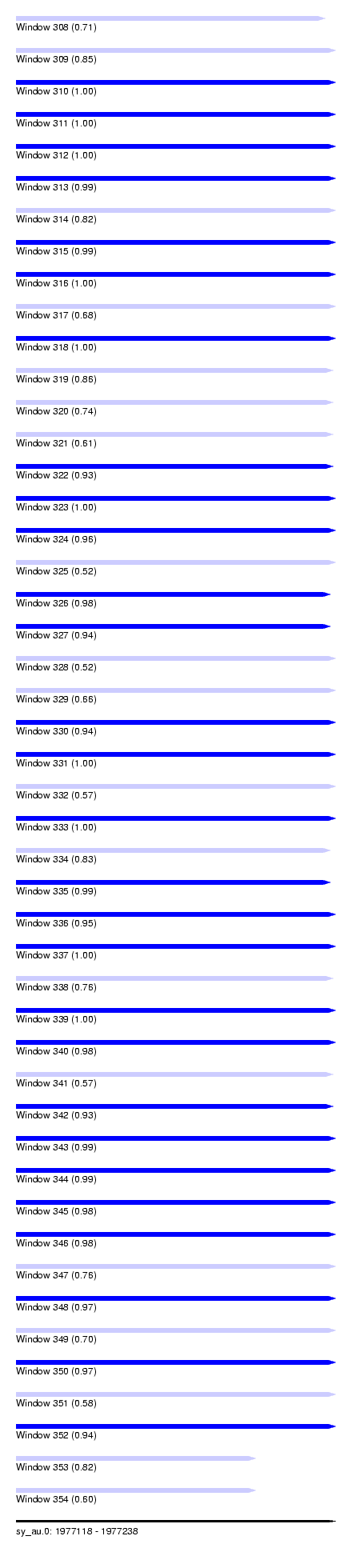
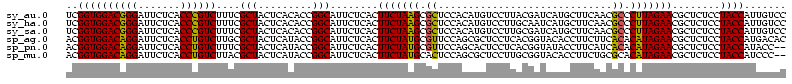
| Sequence ID | sy_au.0 |
|---|---|
| Location | 1,977,118 – 1,977,238 |
| Length | 120 |
| Max. P | 0.999910 |

| Location | 1,977,118 – 1,977,234 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.53 |
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -26.21 |
| Energy contribution | -23.55 |
| Covariance contribution | -2.66 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708828 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 116 + 2809422/0-120 UUUCCC---AACUUCGGUUA-UAAGAUCCCUCAAAGAUGAUGAGGUUAAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUCGAUCGAAGACUUAAUCAAAAU ......---..((((((((.-.......(((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))))))............. ( -32.00) >sy_ha.0 1576954 116 - 2685015/0-120 UUUCCC---AACUUCGGUUA-UAAGAUCCCUCAAAGAUGAUGAGGUUAAUAGGUUCGAGGUGGAAGCGUGGUGACACGUGGAGCUGACGAAUACUAAUCGAUCGAAGACUUAAUCAAUUU ......---..((((((((.-.......(((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))))))............. ( -33.40) >sy_sa.0 1551999 116 - 2516575/0-120 UUUCCC---AACUUCGGUUA-UAAGAUCCCUCAAAGAUGAUGAGGUUAAUAGGUUCGAGGUGGAAGCAUAGCGAUAUGUGCAGCUGACGAAUACUAAUCGAUCGAAGACUUAAUCAAAUU ......---..((((((((.-.......(((((.......)))))....((((((((.(((....(((((......))))).)))..))))).)))...))))))))............. ( -27.00) >sp_ag.0 2133292 120 - 2160267/0-120 UUUCCCAUGAUUUUAUAUCAGUAAGAGCCCUGAGAGAUGAUCAGGUAGAUAGGUUAGGAGUGUAAUUGUGGUGACACAUUUAGCGGACUAAUACUAAUAGCUCGAGGACUUAUCCAAAAA ..((((.((((.....))))....(((((((((.......)))))....((((((((..((.(((.((((....)))).)))))...))))).)))...))))).)))............ ( -30.20) >sp_pn.0 1692470 120 + 2038615/0-120 UUUCCCAUGAUUAUAUAUCAGUAAGAGCCCUGAGAGAUGAUCAGGUAGAUAGGUUAGAAGUGGAAGUGUGGCGACACAUGUAGCGGACUAAUACUAAUAGCUCGAGGACUUAUCCAAAGU ....((.(((((((...((((........))))...)))))))))..((((((((....((....(((((....)))))...))(..(((.......)))..)...))))))))...... ( -30.20) >sp_mu.0 1873490 120 + 2030921/0-120 UUUCCCAUAACGUUCAGUUAGUAAGAGCCCUGAGAGAAGAACAGGUAGAUAGGUUGGGAGUGGAAGCGUUGUGAGACGUGAAGCGGACCAAUACUAAUCGCUCGAGGACUUAUCCAAAAA ..((((.((((.....))))....((((((((.........))))..((((((((((..((....(((((....)))))...))...))))).)).)))))))).)))............ ( -28.30) >consensus UUUCCC___AACUUCGGUUA_UAAGAGCCCUCAAAGAUGAUCAGGUAAAUAGGUUCGAAGUGGAAGCGUGGUGACACGUGGAGCGGACGAAUACUAAUCGAUCGAAGACUUAACCAAAAU ........................(((((((((.......)))))....((((((((..((....(((((....)))))...))...))))).)))...))))................. (-26.21 = -23.55 + -2.66) # Strand winner: reverse (1.00)

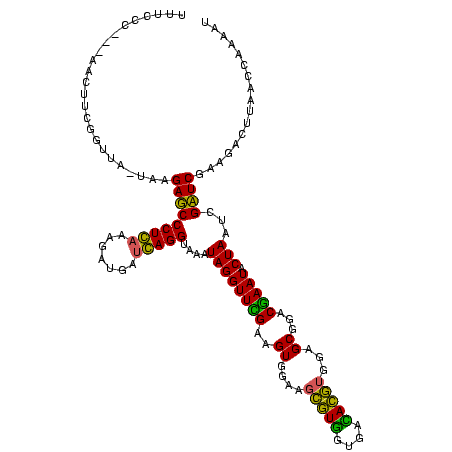
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.67 |
| Mean single sequence MFE | -39.37 |
| Consensus MFE | -38.08 |
| Energy contribution | -35.45 |
| Covariance contribution | -2.63 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/120-240 GACCGGGAUGGACAUACCUCUGGUGUACCAGUUGUCGUGCCAACGGCAUAGCUGGGUAGCUAUGUGUGGACGGGAUAAGUGCUGAAAGCAUCUAAGCAUGAAGCCCCCCUCAAGAUGAGA ....(((..(..(((((...((((...(((((((((((....))))))..)))))...)))).)))))..)(((....(((((...........)))))....))))))........... ( -41.00) >sy_ha.0 1576954 120 - 2685015/120-240 GACCGGGAUGGACAUACCUCUGGUGUACCAGUUGUCGUGCCAACGGCAUAGCUGGGUAGCUAUGUAUGGACGGGAUAAGUGCUGAAAGCAUCUAAGCAUGAAGCCCCCCUCAAGAUGAGA ....(((..(..(((((...((((...(((((((((((....))))))..)))))...)))).)))))..)(((....(((((...........)))))....))))))........... ( -41.00) >sy_sa.0 1551999 120 - 2516575/120-240 GACCGGGAUGGACAUACCUCUGGUGUACCAGUUGUCGUGCCAACGGCAUCGCUGGGUAGCUAUGUAUGGACGGGAUAAGUGCUGAAAGCAUCUAAGCAUGAAGCCCCCCUCAAGAUGAGA ....(((..(..(((((...((((...(((((((((((....))))))..)))))...)))).)))))..)(((....(((((...........)))))....))))))........... ( -41.00) >sp_ag.0 2133292 120 - 2160267/120-240 GACCAGAGUGGACUUACCGCUGGUGUACCAGUUGUCUUGCCAAAGGCAUCGCUGGGUAGCUAUGUAGGGAAGGGAUAAGCGCUGAAAGCAUCUAAGUGCGAAGCCCACCUCAAGAUGAGA ..((..(((((.....)))))(((...(((((((((((....))))))..)))))...))).....))...(((....(((((...........)))))....)))..(((.....))). ( -38.10) >sp_pn.0 1692470 120 + 2038615/120-240 GACCAGAGUGGACUUACCGCUGGUGUACCAGUUGUCUUGCCAAAGGCAUCGCUGGGUAGCUAUGUAGGGAAGGGAUAAACGCUGAAAGCAUCUAAGUGUGAAACCCACCUCAAGAUGAGA ..((..(((((.....)))))(((...(((((((((((....))))))..)))))...))).....))...(((....(((((...........)))))....)))..(((.....))). ( -35.10) >sp_mu.0 1873490 120 + 2030921/120-240 GACCAGAGUGGACUUACCGCUGGUGUACCAGUUGUUCUGCCAAGAGCAUCGCUGGGUAGCUAAGUAGGGAGGGGAUAAACGCUGAAAGCAUCUAAGUGUGAAGCCCCCCUCAAGAUGAGA ..((..(((((.....)))))(((...(((((((((((....))))))..)))))...))).....))((((((....(((((...........)))))......))))))......... ( -40.00) >consensus GACCAGAAUGGACAUACCGCUGGUGUACCAGUUGUCGUGCCAACGGCAUCGCUGGGUAGCUAUGUAGGGACGGGAUAAGCGCUGAAAGCAUCUAAGCAUGAAGCCCCCCUCAAGAUGAGA ..((.....)).(((((...((((...(((((((((((....))))))..)))))...)))).)))))...(((....(((((...........)))))....)))..(((.....))). (-38.08 = -35.45 + -2.63) # Strand winner: reverse (1.00)

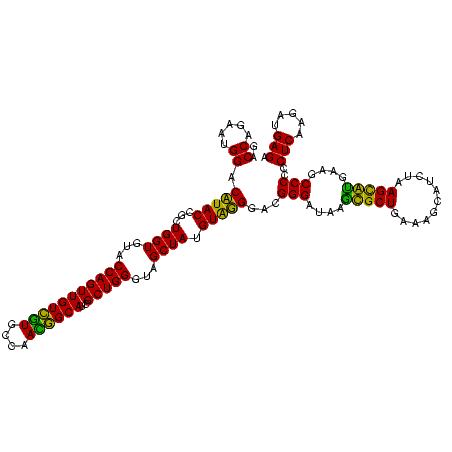
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -54.10 |
| Consensus MFE | -54.00 |
| Energy contribution | -53.50 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.02 |
| Mean z-score | -1.95 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.91 |
| SVM RNA-class probability | 0.999700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/280-400 UGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGUG .(.(((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))).(((..(.(((((((....))).)))))..)))....)).)... ( -52.70) >sy_ha.0 1576954 120 - 2685015/280-400 UGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGUG .(.(((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))).(((..(.(((((((....))).)))))..)))....)).)... ( -52.70) >sy_sa.0 1551999 120 - 2516575/280-400 UGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGUG .(.(((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))).(((..(.(((((((....))).)))))..)))....)).)... ( -52.70) >sp_ag.0 2133292 120 - 2160267/280-400 UGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGCG .........(((((((((((((((((....))))))).)))))(((((.....))))).....))))).((((((.((((..(.(((((((....))).)))).)..))))..).))))) ( -55.50) >sp_pn.0 1692470 120 + 2038615/280-400 UGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGCG .........(((((((((((((((((....))))))).)))))(((((.....))))).....))))).((((((.((((..(.(((((((....))).)))).)..))))..).))))) ( -55.50) >sp_mu.0 1873490 120 + 2030921/280-400 UGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGCG .........(((((((((((((((((....))))))).)))))(((((.....))))).....))))).((((((.((((..(.(((((((....))).)))).)..))))..).))))) ( -55.50) >consensus UGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGCG .(.(((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))).(((..(.(((((((....))).)))))..)))....)).)... (-54.00 = -53.50 + -0.50) # Strand winner: reverse (1.00)

| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -46.30 |
| Consensus MFE | -46.60 |
| Energy contribution | -46.10 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.41 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/320-440 CCCAGCUCGCGUACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGAACUCUUGGGG (((((((((((((.........(((((.....)))))...((((((.(........).)))))).)))).)))))((((...(((((.....)))))...))))...........)))). ( -45.80) >sy_ha.0 1576954 120 - 2685015/320-440 CCCAGCUCGCGUACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGAACUCUUGGGG (((((((((((((.........(((((.....)))))...((((((.(........).)))))).)))).)))))((((...(((((.....)))))...))))...........)))). ( -45.80) >sy_sa.0 1551999 120 - 2516575/320-440 CCCAGCUCGCGUACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGAACUCUUGGGG (((((((((((((.........(((((.....)))))...((((((.(........).)))))).)))).)))))((((...(((((.....)))))...))))...........)))). ( -45.80) >sp_ag.0 2133292 120 - 2160267/320-440 CCCAGCUCGCGUGCCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGACGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGAACUCUUGGGG ((((((((((((..........(((((.....)))))...((((((.(........).))))))..))).)))))((((...(((((.....)))))...))))...........)))). ( -46.80) >sp_pn.0 1692470 120 + 2038615/320-440 CCCAGCUCGCGUGCCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGACGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGAACUCUUGGGG ((((((((((((..........(((((.....)))))...((((((.(........).))))))..))).)))))((((...(((((.....)))))...))))...........)))). ( -46.80) >sp_mu.0 1873490 120 + 2030921/320-440 CCCAGCUCGCGUGCCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGACGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGAACUCUUGGGG ((((((((((((..........(((((.....)))))...((((((.(........).))))))..))).)))))((((...(((((.....)))))...))))...........)))). ( -46.80) >consensus CCCAGCUCGCGUACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGACGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGAACUCUUGGGG (((((((((((((.........(((((.....)))))...((((((.(........).)))))).)))).)))))((((...(((((.....)))))...))))...........)))). (-46.60 = -46.10 + -0.50)

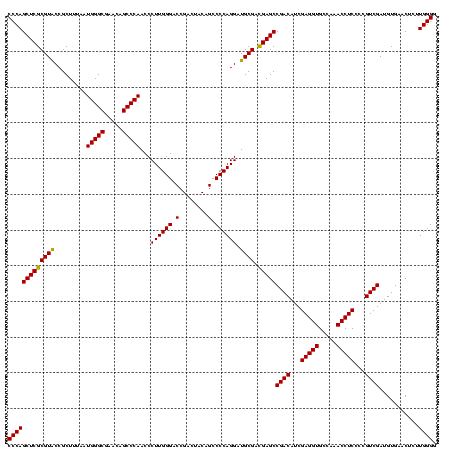
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -56.45 |
| Consensus MFE | -56.95 |
| Energy contribution | -56.45 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.02 |
| Mean z-score | -2.20 |
| Structure conservation index | 1.01 |
| SVM decision value | 4.50 |
| SVM RNA-class probability | 0.999910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/320-440 CCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGG .((((...((((....(((((..(((((.....)))))..)))))((..(((((((((((((((((....))))))).)))))(((((.....))))).....)))))..)))))))))) ( -55.70) >sy_ha.0 1576954 120 - 2685015/320-440 CCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGG .((((...((((....(((((..(((((.....)))))..)))))((..(((((((((((((((((....))))))).)))))(((((.....))))).....)))))..)))))))))) ( -55.70) >sy_sa.0 1551999 120 - 2516575/320-440 CCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGG .((((...((((....(((((..(((((.....)))))..)))))((..(((((((((((((((((....))))))).)))))(((((.....))))).....)))))..)))))))))) ( -55.70) >sp_ag.0 2133292 120 - 2160267/320-440 CCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGG .((((...((((....(((((..(((((.....)))))..)))))((..(((((((((((((((((....))))))).)))))(((((.....))))).....)))))..)))))))))) ( -57.20) >sp_pn.0 1692470 120 + 2038615/320-440 CCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGG .((((...((((....(((((..(((((.....)))))..)))))((..(((((((((((((((((....))))))).)))))(((((.....))))).....)))))..)))))))))) ( -57.20) >sp_mu.0 1873490 120 + 2030921/320-440 CCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGG .((((...((((....(((((..(((((.....)))))..)))))((..(((((((((((((((((....))))))).)))))(((((.....))))).....)))))..)))))))))) ( -57.20) >consensus CCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGG .((((...((((....(((((..(((((.....)))))..)))))((..(((((((((((((((((....))))))).)))))(((((.....))))).....)))))..)))))))))) (-56.95 = -56.45 + -0.50) # Strand winner: reverse (1.00)

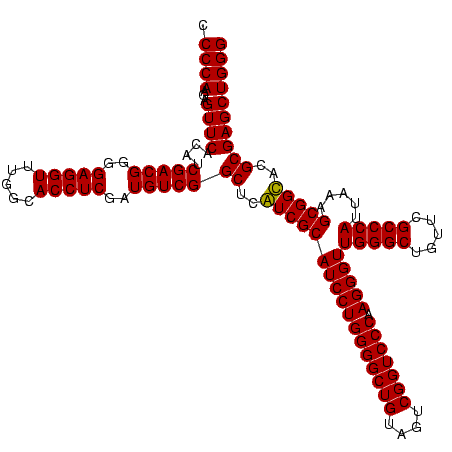
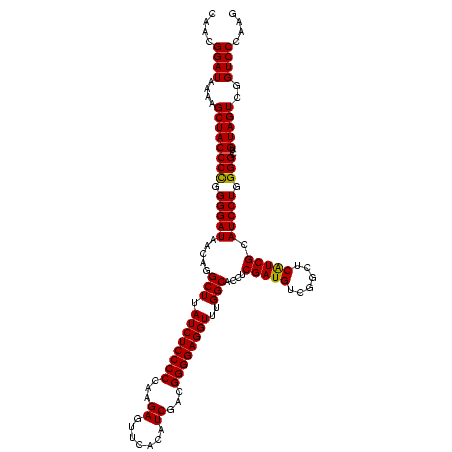
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.00 |
| Mean single sequence MFE | -44.60 |
| Consensus MFE | -45.10 |
| Energy contribution | -44.60 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.29 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992280 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/360-480 CAACGGAUAAAAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAG ....((((....((((((((.(((((.....(((.((((((((...((.......))...))))))))..)))....(((((......))))).))))).)))..)))))..)))).... ( -45.70) >sy_ha.0 1576954 120 - 2685015/360-480 CAACGGAUAAAAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAG ....((((....((((((((.(((((.....(((.((((((((...((.......))...))))))))..)))....(((((......))))).))))).)))..)))))..)))).... ( -45.70) >sy_sa.0 1551999 120 - 2516575/360-480 CAACGGAUAAAAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAG ....((((....((((((((.(((((.....(((.((((((((...((.......))...))))))))..)))....(((((......))))).))))).)))..)))))..)))).... ( -45.70) >sp_ag.0 2133292 120 - 2160267/360-480 CAACGGAUAAAAGCUACCCUGGGGAUAACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAG ....((((....((((((((.(((((.....(((.((((((((...((.......))...))))))))..)))....(((((......))))).))))).)))..)))))..)))).... ( -43.50) >sp_pn.0 1692470 120 + 2038615/360-480 CAACGGAUAAAAGCUACCCUGGGGAUAACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAG ....((((....((((((((.(((((.....(((.((((((((...((.......))...))))))))..)))....(((((......))))).))))).)))..)))))..)))).... ( -43.50) >sp_mu.0 1873490 120 + 2030921/360-480 CAACGGAUAAAAGCUACCCUGGGGAUAACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAG ....((((....((((((((.(((((.....(((.((((((((...((.......))...))))))))..)))....(((((......))))).))))).)))..)))))..)))).... ( -43.50) >consensus CAACGGAUAAAAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAG ....((((....((((((((.(((((.....(((.((((((((...((.......))...))))))))..)))....(((((......))))).))))).)))..)))))..)))).... (-45.10 = -44.60 + -0.50) # Strand winner: reverse (0.95)

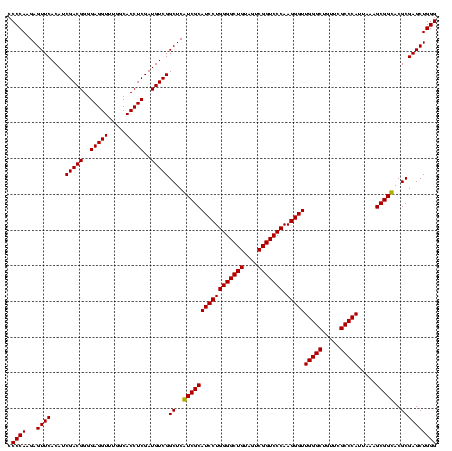
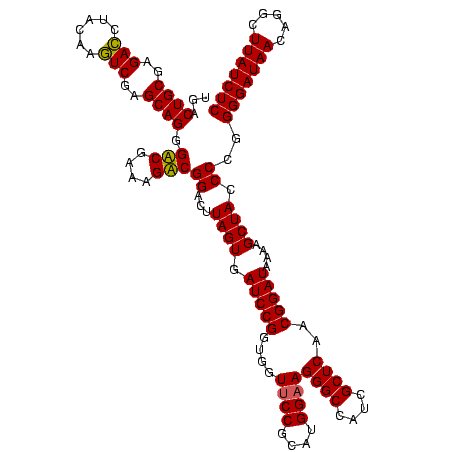
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -40.50 |
| Consensus MFE | -38.33 |
| Energy contribution | -37.55 |
| Covariance contribution | -0.78 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/440-560 UGACUGCGAGACCUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAAAAGCUACCCCGGGGAUAACAGGCUUAUCUC ...((((..(((......)))..)))).(((....)))((...((((.((((((((....)))......((((....))))..)))))....))))..)).(((((((.....))))))) ( -39.30) >sy_ha.0 1576954 120 - 2685015/440-560 UGACUGCGAGACCUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAAAAGCUACCCCGGGGAUAACAGGCUUAUCUC ...((((..(((......)))..)))).(((....)))((...((((.((((((((....)))......((((....))))..)))))....))))..)).(((((((.....))))))) ( -39.30) >sy_sa.0 1551999 120 - 2516575/440-560 UGACUGCGAGACUUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAAAAGCUACCCCGGGGAUAACAGGCUUAUCUC ...((((..((((....))))..)))).(((....)))((...((((.((((((((....)))......((((....))))..)))))....))))..)).(((((((.....))))))) ( -40.50) >sp_ag.0 2133292 120 - 2160267/440-560 UGACUGCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGCUACCCUGGGGAUAACAGGCUUAUCUC ...((((..(((......)))..)))).(((....)))(((..((((.(((((....((((....))))((((....))))..)))))....)))))))..(((((((.....))))))) ( -41.50) >sp_pn.0 1692470 120 + 2038615/440-560 UGACUGCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGCUACCCUGGGGAUAACAGGCUUAUCUC ...((((..(((......)))..)))).(((....)))(((..((((.(((((....((((....))))((((....))))..)))))....)))))))..(((((((.....))))))) ( -41.50) >sp_mu.0 1873490 120 + 2030921/440-560 UGACUGCGAGACAGACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUACCGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGCUACCCUGGGGAUAACAGGCUUAUCUC ...((((..(((......)))..)))).(((....)))(((..((((.((((((((((.((........))))))))(.....)))))....)))))))..(((((((.....))))))) ( -40.90) >consensus UGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGACGGACUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAAAAGCUACCCCGGGGAUAACAGGCUUAUCUC ...((((..(((......)))..)))).(((....)))((...((((.(((((....((((....))))((((....))))..)))))....)))).))..(((((((.....))))))) (-38.33 = -37.55 + -0.78) # Strand winner: reverse (1.00)


| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -37.85 |
| Consensus MFE | -37.98 |
| Energy contribution | -36.32 |
| Covariance contribution | -1.66 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.23 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987737 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/480-600 GUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCU ...(((......((((.((((.(..((((........))))..((((..(((......)))..)))).(((....)))).)))).)))))))((((((.((........))))))))... ( -36.80) >sy_ha.0 1576954 120 - 2685015/480-600 GUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCU ...(((......((((.((((.(..((((........))))..((((..(((......)))..)))).(((....)))).)))).)))))))((((((.((........))))))))... ( -36.80) >sy_sa.0 1551999 120 - 2516575/480-600 GUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGAAGCUUGACUGCGAGACUUACAAGUCGAGCAGGGUCGAAAGACGGACUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCU ...(((......((((.((((.(..((((........))))..((((..((((....))))..)))).(((....)))).)))).)))))))((((((.((........))))))))... ( -38.00) >sp_ag.0 2133292 120 - 2160267/480-600 GUUGGAAAUCAGUCGUAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGUAUGGAAGGGCCAUCGCU ...(((..(((......(((((((..(((.((((....)))))))((....))))).))))((((...(((....)))..))))..))))))((((((.((........))))))))... ( -37.70) >sp_pn.0 1692470 120 + 2038615/480-600 GUUGGAAAUCAUUCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGUAUGGAAGGGCCAUCGCU ...(((......((((.(((((((..(((.((((....)))))))((....))))).))))((((...(((....)))..)))).)))))))((((((.((........))))))))... ( -39.30) >sp_mu.0 1873490 120 + 2030921/480-600 GUUGGAAAUCAGUCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGACAGACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUACCGUAUGGAAGGGCCAUCGCU ...........(((((.((((.(..............((((((((.(......)...))))))))...(((....)))).)))).))))).(((((((.((........))))))))).. ( -38.50) >consensus GUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGACGGACUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCU ...(((......((((.((((.(.......((((....)))).((((..(((......)))..)))).(((....)))).)))).)))))))((((((.((........))))))))... (-37.98 = -36.32 + -1.66) # Strand winner: reverse (1.00)
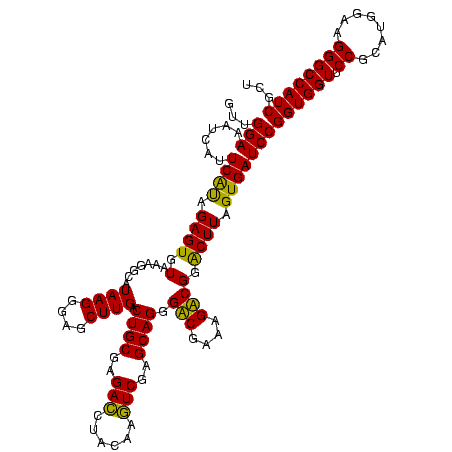
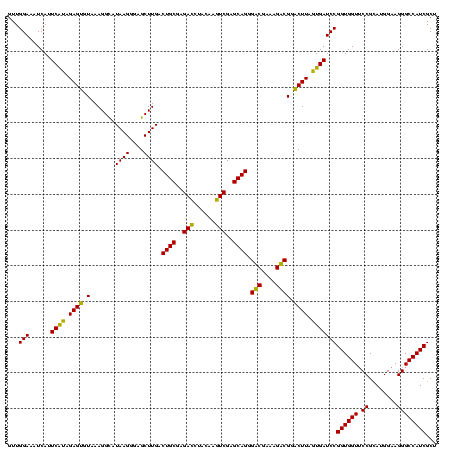
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.00 |
| Mean single sequence MFE | -37.45 |
| Consensus MFE | -36.98 |
| Energy contribution | -34.82 |
| Covariance contribution | -2.16 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.30 |
| SVM RNA-class probability | 0.999863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/520-640 CUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGUCGAAAGACGG .............(((...)))..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....))).. ( -36.90) >sy_ha.0 1576954 120 - 2685015/520-640 CUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGUCGAAAGACGG .............(((...)))..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....))).. ( -36.90) >sy_sa.0 1551999 120 - 2516575/520-640 CUAAAGAGUAACGGAGGCGCUCAAAGGUUUCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGAAGCUUGACUGCGAGACUUACAAGUCGAGCAGGGUCGAAAGACGG .....((((..(...)..))))..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..((((....))))..)))).(((....))).. ( -38.30) >sp_ag.0 2133292 120 - 2160267/520-640 CUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGGAAAUCAGUCGUAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGG ............((......))..(((((((((..(((((((.....)))))))......((((....)))))))))))))..((((..(((......)))..)))).(((....))).. ( -37.50) >sp_pn.0 1692470 120 + 2038615/520-640 CUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGG ............((......))..(((((((((..(((((((.....)))))))((.....)).........)))))))))..((((..(((......)))..)))).(((....))).. ( -35.50) >sp_mu.0 1873490 120 + 2030921/520-640 CUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGGAAAUCAGUCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGACAGACAAGUCGAGCAGGGACGAAAGUCGG .....((((.......))))((..(((((((((..(((((((.....)))))))((.....)).........)))))))))..((((..(((......)))..)))).(((....))))) ( -39.60) >consensus CUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGACGG ............((......))..(((((((((..(((((((.....))))))).....(((......))).)))))))))..((((..(((......)))..)))).(((....))).. (-36.98 = -34.82 + -2.16) # Strand winner: reverse (1.00)

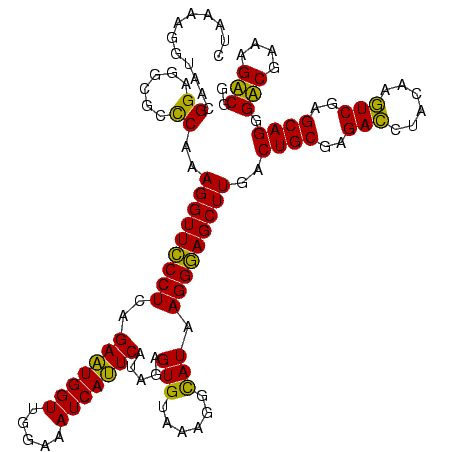
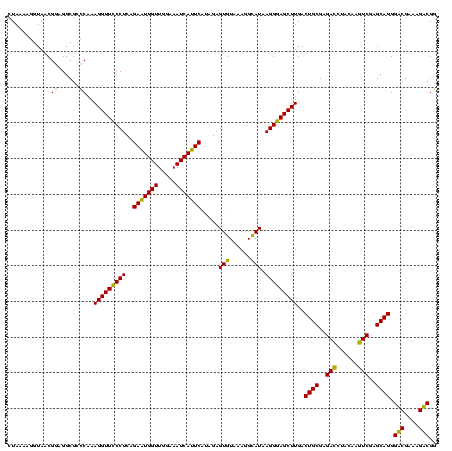
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -30.77 |
| Consensus MFE | -27.05 |
| Energy contribution | -26.33 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.676432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/560-680 AGCUCCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGCCUGACACUGUC .(((((((((((......))......(((((.........))))))))))))......(..(((((((...........)))))))..)))....((((..(.....)..))))...... ( -28.70) >sy_ha.0 1576954 120 - 2685015/560-680 AGCUCCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGCCUGACACUGUC .(((((((((((......))......(((((.........))))))))))))......(..(((((((...........)))))))..)))....((((..(.....)..))))...... ( -28.70) >sy_sa.0 1551999 120 - 2516575/560-680 AGCUUCCUUAUGCCUUUACACUCUAUGAAUGAUUUCCAACCAUUCUGAGGAAACCUUUGAGCGCCUCCGUUACUCUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCAUCUGACACUGUC .(((((((((((......))......(((((.........))))))))))))......(..(((((((...........)))))))..)))....((((...........))))...... ( -26.10) >sp_ag.0 2133292 120 - 2160267/560-680 AGCUCCCUUAUACCUUUACACUCUACGACUGAUUUCCAACCAGUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUC ...(((((((................(((((.........))))))))))))......((((((((((...........))))))(((.......))).....))))............. ( -34.39) >sp_pn.0 1692470 120 + 2038615/560-680 AGCUCCCUUAUACCUUUACACUCUGCGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUC ...(((((((........(.....).(((((.........))))))))))))......((((((((((...........))))))(((.......))).....))))............. ( -32.20) >sp_mu.0 1873490 120 + 2030921/560-680 AGCUCCCUUAUACCUUUACACUCUGCGACUGAUUUCCAACCAGUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUC ...(((((((........(.....).(((((.........))))))))))))......((((((((((...........))))))(((.......))).....))))............. ( -34.50) >consensus AGCUCCCUUAUACCUUUACACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUC .(((((((((................(((((.........)))))))))))).........(((((((...........)))))))...))....(((.............)))...... (-27.05 = -26.33 + -0.72)
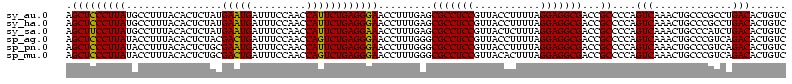
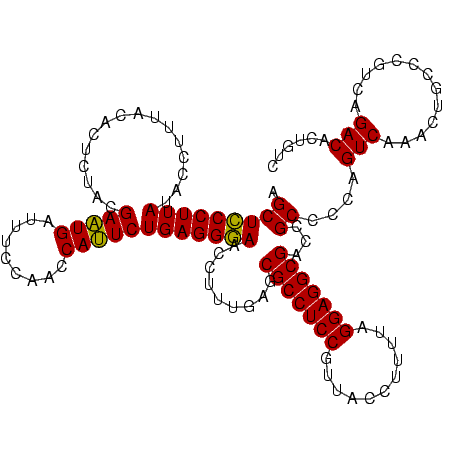
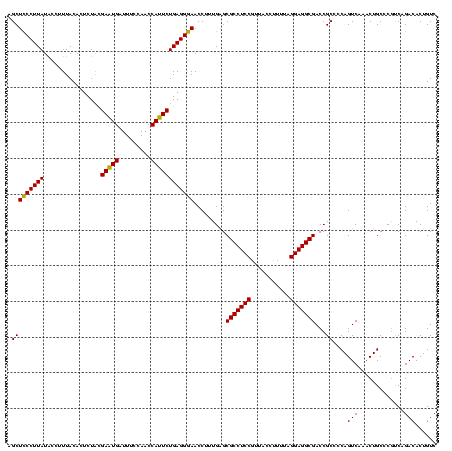
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -41.93 |
| Consensus MFE | -41.94 |
| Energy contribution | -40.72 |
| Covariance contribution | -1.22 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.89 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.98 |
| SVM RNA-class probability | 0.999739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
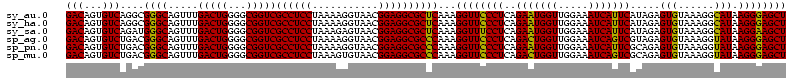
>sy_au.0 1977118 120 + 2809422/560-680 GACAGUGUCAGGCGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCU ..(...(((((((.....)))))))...)..(..(((((((...........)))))))..)...((((((((..(((((((.....))))))).....((((....)))).)))))))) ( -41.60) >sy_ha.0 1576954 120 - 2685015/560-680 GACAGUGUCAGGCGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCUCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCU ..(...(((((((.....)))))))...)..(..(((((((...........)))))))..)...((((((((..(((((((.....))))))).....((((....)))).)))))))) ( -41.60) >sy_sa.0 1551999 120 - 2516575/560-680 GACAGUGUCAGAUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGAGUAACGGAGGCGCUCAAAGGUUUCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGAAGCU (((...)))...(((((.....(((((...)))))((((((...........)))))))))))..((((((((..(((((((.....))))))).....((((....)))).)))))))) ( -39.60) >sp_ag.0 2133292 120 - 2160267/560-680 GACAGUGUCUGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGGAAAUCAGUCGUAGAGUGUAAAGGUAUAAGGGAGCU (((...)))....((((.....(((((...)))))((((((...........))))))))))...((((((((..(((((((.....)))))))......((((....)))))))))))) ( -43.50) >sp_pn.0 1692470 120 + 2038615/560-680 GACAGUGUCUGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGUAUAAGGGAGCU (((...)))....((((.....(((((...)))))((((((...........))))))))))...((((((((..(((((((.....)))))))((.....)).........)))))))) ( -41.50) >sp_mu.0 1873490 120 + 2030921/560-680 GACAGUGUCUGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGGAAAUCAGUCGCAGAGUGUAAAGGUAUAAGGGAGCU (((...)))....((((.....(((((...)))))((((((...........))))))))))...((((((((..(((((((.....)))))))((.....)).........)))))))) ( -43.80) >consensus GACAGUGUCAGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCAUAGAGUGUAAAGGCAUAAGGGAGCU (((...)))....((((.....(((((...)))))((((((...........))))))))))...((((((((..(((((((.....))))))).....(((......))).)))))))) (-41.94 = -40.72 + -1.22) # Strand winner: reverse (1.00)
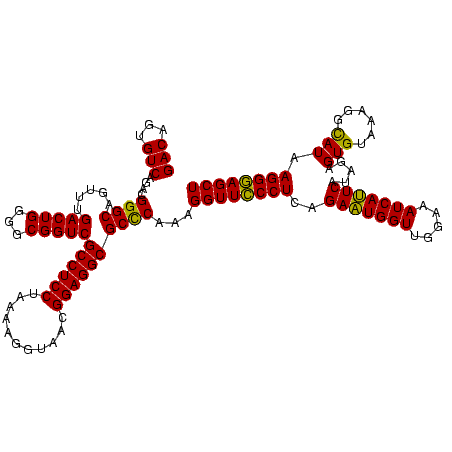
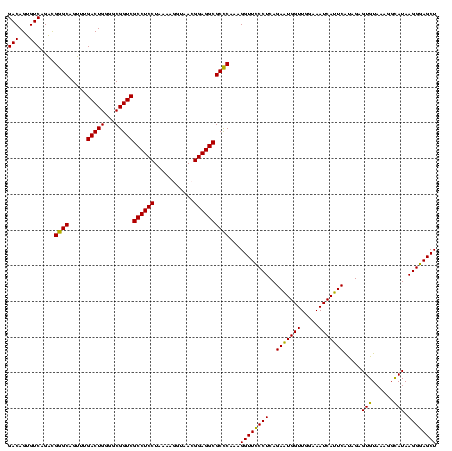
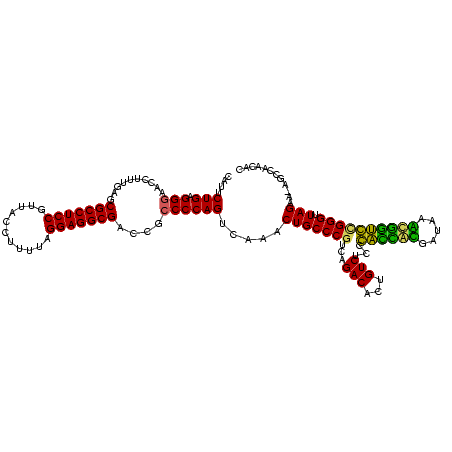
| Location | 1,977,118 – 1,977,237 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.48 |
| Mean single sequence MFE | -40.85 |
| Consensus MFE | -36.59 |
| Energy contribution | -33.92 |
| Covariance contribution | -2.66 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 119 + 2809422/600-720 CAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAA-AGCCAACAC ....(((.(((......((..(((((((...........)))))))..)))))))).....((((((((..(((...)))....(((((.....)))))))))).)))..-......... ( -39.20) >sy_ha.0 1576954 119 - 2685015/600-720 CAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAA-AGCCAACAC ....(((.(((......((..(((((((...........)))))))..)))))))).....((((((((..(((...)))....(((((.....)))))))))).)))..-......... ( -39.20) >sy_sa.0 1551999 119 - 2516575/600-720 CAUUCUGAGGAAACCUUUGAGCGCCUCCGUUACUCUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCAUCUGACACUGUCUCCCACCAUGAUAAAUGGUGCGGGUUAGAA-AUCCAACAC ........(((......((.((((((((...........))))))(((.......))).....)).))((((((.(((.....((((((.....))))))))))))))).-.)))..... ( -35.20) >sp_ag.0 2133292 120 - 2160267/600-720 CAGUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCGAUAGGGAUUGCCUAUCUGGGUUAGAGUAGCCAUAAC ..(((((((((.......((((((((((...........))))))....))))(((.....)))))).)))))).(((.(((.((((((.....)))))).))).)))............ ( -46.00) >sp_pn.0 1692470 120 + 2038615/600-720 CAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCGAUAGGGAUCACCUAUCUGGGUUAGAGUGGCCAUAAC ...((((((((.......((((((((((...........))))))....))))(((.....)))))).)))))((((.((((.((((((.....)))))).)))..).))))........ ( -44.80) >sp_mu.0 1873490 120 + 2030921/600-720 CAGUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCGUAGAUGAUCAACCUACCGGGUUAGAGUAGCCAUCAC ..(((((((((.......((((((((((...........))))))....))))(((.....)))))).)))))).(((.(((.((((.........)))).))).)))............ ( -40.70) >consensus CAUUCUGAGGGAACCUUUGAGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCCACCACGAUAAACGGUCCGGGUUAGAA_AGCCAACAC ....(((.(((..........(((((((...........)))))))....)))))).....(((((((...(((...)))...((((((.....)))))))))).)))............ (-36.59 = -33.92 + -2.66)

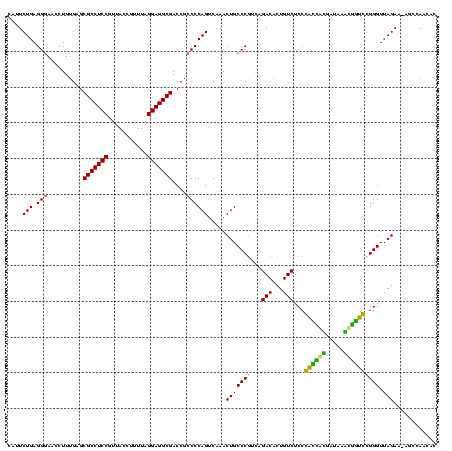
| Location | 1,977,118 – 1,977,237 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.22 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -36.55 |
| Energy contribution | -33.58 |
| Covariance contribution | -2.97 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.739374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 119 + 2809422/640-760 GAGGCGACCGCCCCAGUCAAACUGCCCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAA-AGCCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAG ((((((((.......)))..(((((((..(((((.(((.....((((((.....))))))))))))))..-(((.......))).)))))))...........)))))............ ( -39.90) >sy_ha.0 1576954 119 - 2685015/640-760 GAGGCGACCGCCCCAGUCAAACUGCCCGCCUGACACUGUCUCCCACCACGAUAAGUGGUGCGGGUUAGAA-AGCCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAG ((((((((.......)))..(((((((..(((((.(((.....((((((.....))))))))))))))..-(((.......))).)))))))...........)))))............ ( -39.90) >sy_sa.0 1551999 119 - 2516575/640-760 GAGGCGACCGCCCCAGUCAAACUGCCCAUCUGACACUGUCUCCCACCAUGAUAAAUGGUGCGGGUUAGAA-AUCCAACACAGCGAGGGUAGUAUCCCACCAACGCCUCCACGUAAGCUAG ((((((((.......)))..(((((((.((((((.(((.....((((((.....))))))))))))))).-.(((......).)))))))))...........)))))............ ( -37.10) >sp_ag.0 2133292 119 - 2160267/640-760 GAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCGAUAGGGAUUGCCUAUCUGGGUUAGAGUAGCCAUAACA-CAAGGGUAGUAUCCCAACAACGCCUCAAACGAAACUGG ((((((((.......)))..(((((((........(((.(((.((((((.....)))))).))).))).((.......)).-...)))))))...........)))))............ ( -35.70) >sp_pn.0 1692470 119 + 2038615/640-760 GAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCGAUAGGGAUCACCUAUCUGGGUUAGAGUGGCCAUAACA-CAAGGGUAGUAUCCCAACAGCGUCUCCUUCGAAACUGG ..(((....)))(((((...(((((((........(((.(((.((((((.....)))))).))).))).(((.......))-)..)))))))..........((.......))..))))) ( -37.40) >sp_mu.0 1873490 119 + 2030921/640-760 GAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCGUAGAUGAUCAACCUACCGGGUUAGAGUAGCCAUCACA-CAAGGGUAGUAUCCCAACAACGCCUCCAAGUAAACUAG ((((((((.......)))..(((((((...((((...))))..((.((((.((((((.....)))).)).....)))))).-...)))))))...........)))))............ ( -32.00) >consensus GAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCCACCACGAUAAACGGUCCGGGUUAGAA_AGCCAACACA_CAAGGGUAGUAUCCCAACAACGCCUCCACGUAAACUAG ((((((((.......)))..(((((((....(((...)))(((((((((.....)))))).))).....................)))))))...........)))))............ (-36.55 = -33.58 + -2.97)

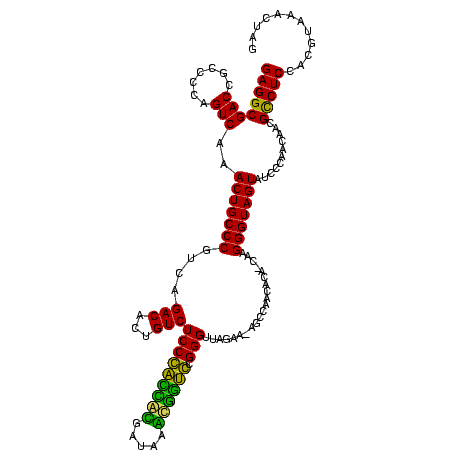
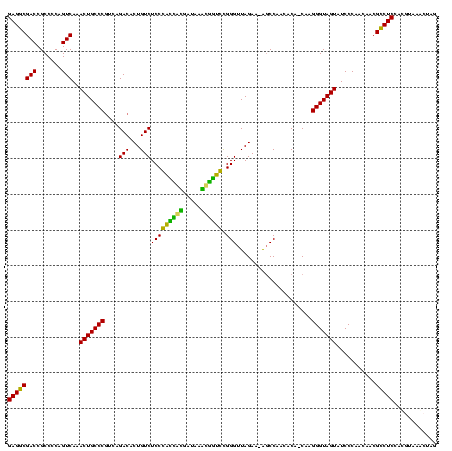
| Location | 1,977,118 – 1,977,237 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.81 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -31.04 |
| Energy contribution | -26.55 |
| Covariance contribution | -4.49 |
| Combinations/Pair | 1.59 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.607999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 119 + 2809422/680-800 UCCCACCACGAUAAGUGGUGCGGGUUAGAA-AGCCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAGCGCUCACGUUUCAAAGGCUCCUACCUAUCCUGUACAAGCU (((((((((.....)))))).)))......-(((....((((...((((((..........(.((((..((((.(((....))).)))).....)))).)))))))...))))....))) ( -35.10) >sy_ha.0 1576954 119 - 2685015/680-800 UCCCACCACGAUAAGUGGUGCGGGUUAGAA-AGCCAACACAGCUAGGGUAGUAUCCCACCAGCGCCUCCACGUAAGCUAGCGCUCACGUUUCAAAGGCUCCUACCUAUCCUGUACAAGCU (((((((((.....)))))).)))......-(((....((((...((((((..........(.((((..((((.(((....))).)))).....)))).)))))))...))))....))) ( -35.10) >sy_sa.0 1551999 119 - 2516575/680-800 UCCCACCAUGAUAAAUGGUGCGGGUUAGAA-AUCCAACACAGCGAGGGUAGUAUCCCACCAACGCCUCCACGUAAGCUAGCGCUCACGUAUCUCAGGCUCCUACCUAUCCUGUACAAGCU (((((((((.....)))))).)))......-.......((((.(((((......)))......((((..((((.(((....))).)))).....)))).........))))))....... ( -32.00) >sp_ag.0 2133292 119 - 2160267/680-800 UCCGAUAGGGAUUGCCUAUCUGGGUUAGAGUAGCCAUAACA-CAAGGGUAGUAUCCCAACAACGCCUCAAACGAAACUGGCGUCCCGUUAUCAUAGGCUCCUACCUAUCCUGUACAUGUG ...((((((....((((((...(((((...)))))(((((.-...(((......)))....(((((............)))))...))))).)))))).....))))))........... ( -33.80) >sp_pn.0 1692470 119 + 2038615/680-800 UCCGAUAGGGAUCACCUAUCUGGGUUAGAGUGGCCAUAACA-CAAGGGUAGUAUCCCAACAGCGUCUCCUUCGAAACUGGCGUCCCGAUCUCUUAGACUCCUACCUAUCCUGUACAUGUG ...((((((.....))))))(((((..((((..........-...(((......)))....(((((............))))).............))))..)))))............. ( -27.60) >sp_mu.0 1873490 119 + 2030921/680-800 UCCGUAGAUGAUCAACCUACCGGGUUAGAGUAGCCAUCACA-CAAGGGUAGUAUCCCAACAACGCCUCCAAGUAAACUAGCGUUCACUCUUCCUUGGCUCCUACCUAUCCUGUACAUGUG ...((((.........))))((((.(((((.(((((.....-...(((......)))...(((((..............)))))..........))))).))..))).))))........ ( -23.74) >consensus UCCCACCACGAUAAACGGUCCGGGUUAGAA_AGCCAACACA_CAAGGGUAGUAUCCCAACAACGCCUCCACGUAAACUAGCGCUCACGUUUCAUAGGCUCCUACCUAUCCUGUACAAGCG ...((((((.....))))))((((.(((.................(((......)))......((((..((((.(((....))).)))).....))))......))).))))........ (-31.04 = -26.55 + -4.49)

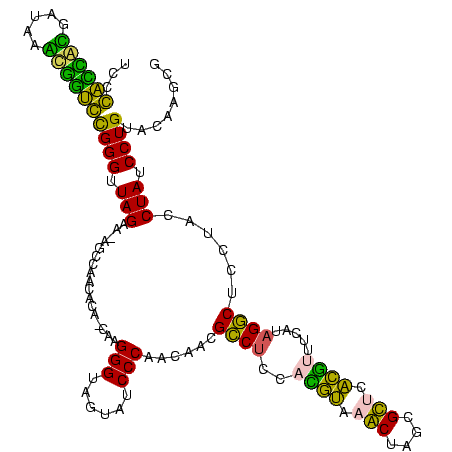
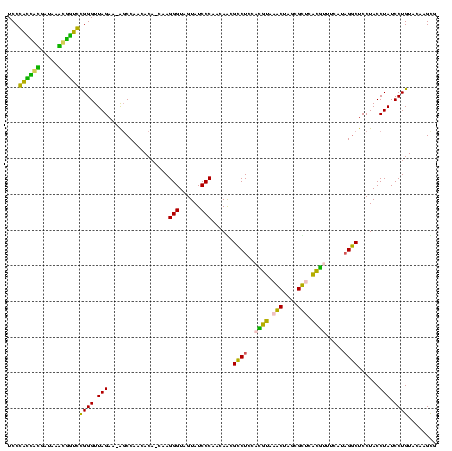
| Location | 1,977,118 – 1,977,237 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.81 |
| Mean single sequence MFE | -41.88 |
| Consensus MFE | -38.17 |
| Energy contribution | -32.18 |
| Covariance contribution | -5.99 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 119 + 2809422/680-800 AGCUUGUACAGGAUAGGUAGGAGCCUUUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCU-UUCUAACCCGCACCACUUAUCGUGGUGGGA ((((..(((((..(((...((.(((((....((((((((....)))))))).))))).))(((((....)))))))).)))))..))))-.......((.((((((.....)))))))). ( -50.60) >sy_ha.0 1576954 119 - 2685015/680-800 AGCUUGUACAGGAUAGGUAGGAGCCUUUGAAACGUGAGCGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUAGCUGUGUUGGCU-UUCUAACCCGCACCACUUAUCGUGGUGGGA ((((..(((((..(((...((.(((((....((((((((....)))))))).))))).))(((((....)))))))).)))))..))))-.......((.((((((.....)))))))). ( -50.60) >sy_sa.0 1551999 119 - 2516575/680-800 AGCUUGUACAGGAUAGGUAGGAGCCUGAGAUACGUGAGCGCUAGCUUACGUGGAGGCGUUGGUGGGAUACUACCCUCGCUGUGUUGGAU-UUCUAACCCGCACCAUUUAUCAUGGUGGGA .((((.(((.......))).))))......(((((((((....)))))))))..((((..(((((....)))))..))))..(((((..-..)))))((.((((((.....)))))))). ( -46.60) >sp_ag.0 2133292 119 - 2160267/680-800 CACAUGUACAGGAUAGGUAGGAGCCUAUGAUAACGGGACGCCAGUUUCGUUUGAGGCGUUGUUGGGAUACUACCCUUG-UGUUAUGGCUACUCUAACCCAGAUAGGCAAUCCCUAUCGGA ..........((.(((((((..((((.....((((..((....))..))))..))))......(((......)))...-........)))).))).))..((((((.....))))))... ( -34.50) >sp_pn.0 1692470 119 + 2038615/680-800 CACAUGUACAGGAUAGGUAGGAGUCUAAGAGAUCGGGACGCCAGUUUCGAAGGAGACGCUGUUGGGAUACUACCCUUG-UGUUAUGGCCACUCUAACCCAGAUAGGUGAUCCCUAUCGGA ..((((.(((.((..(((((...(((((.((.(((..((....))..))).(....).)).)))))...))))).)).-)))))))...........((.((((((.....)))))))). ( -37.60) >sp_mu.0 1873490 119 + 2030921/680-800 CACAUGUACAGGAUAGGUAGGAGCCAAGGAAGAGUGAACGCUAGUUUACUUGGAGGCGUUGUUGGGAUACUACCCUUG-UGUGAUGGCUACUCUAACCCGGUAGGUUGAUCAUCUACGGA ...............(((.((((........((((((((....))))))))..((.(((..(.(((......)))...-.)..))).)).)))).)))(.((((((.....)))))).). ( -31.40) >consensus AACAUGUACAGGAUAGGUAGGAGCCUAUGAAACGUGAACGCUAGCUUACGUGGAGGCGCUGGUGGGAUACUACCCUUG_UGUGAUGGCU_CUCUAACCCGCACCACUUAUCAUGGUCGGA ...............(((.(((((((.....((((((((....))))))))..))))((..(.(((......)))........)..))...))).)))(.((((((.....)))))).). (-38.17 = -32.18 + -5.99) # Strand winner: reverse (1.00)

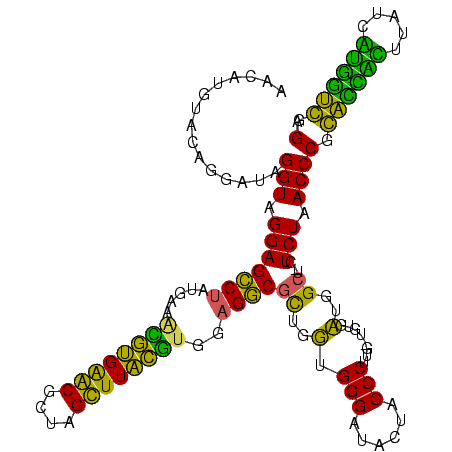
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -39.35 |
| Consensus MFE | -38.00 |
| Energy contribution | -38.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.998782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/840-960 GGUCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUGAUUUCACCGAGUCUCUCGUUGAGACAGUGCCCAAAUCGUUACGCCUUUCGUGCGGGUCGGAACUUACCCGAC (((..(((((.((((.((((((((.((((........))))))))((((((((((...(((((.....))))).)))...))))))).........)))))))).)))))...))).... ( -40.70) >sy_ha.0 1576954 120 - 2685015/840-960 GGUCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUGAUUUCACCGAGUCUCUCGUUGAGACAGUGCCCAAAUCGUUACGCCUUUCGUGCGGGUCGGAACUUACCCGAC (((..(((((.((((.((((((((.((((........))))))))((((((((((...(((((.....))))).)))...))))))).........)))))))).)))))...))).... ( -40.70) >sy_sa.0 1551999 120 - 2516575/840-960 GGUCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUGAUUUCACCGAGUCUCUCGUUGAGACAGUGCCCAAAUCGUUACGCCUUUCGUGCGGGUCGGAACUUACCCGAC (((..(((((.((((.((((((((.((((........))))))))((((((((((...(((((.....))))).)))...))))))).........)))))))).)))))...))).... ( -40.70) >sp_ag.0 2133292 120 - 2160267/840-960 GGUCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAAAAUUUCACCGAGUCUCUCGUUGAGACAGUGCCCAAAUCAUUACGCCUUUCGUGCGGGUCGGAACUUACCCGAC (((..(((((.((((.(((((((((((((........)))))................(((((.....)))))..)))))..........(.....)))))))).)))))...))).... ( -38.00) >sp_pn.0 1692470 120 + 2038615/840-960 GGUCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAAAAUUUCACCGAGUCUCUCGUUGAGACAGUGCCCAAAUCAUUACGCCUUUCGUGCGGGUCGGAACUUACCCGAC (((..(((((.((((.(((((((((((((........)))))................(((((.....)))))..)))))..........(.....)))))))).)))))...))).... ( -38.00) >sp_mu.0 1873490 120 + 2030921/840-960 GGUCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAAAAUUUCACCGAGUCUCUCGUUGAGACAGUGCCCAAAUCAUUACGCCUUUCGUGCGGGUCGGAACUUACCCGAC (((..(((((.((((.(((((((((((((........)))))................(((((.....)))))..)))))..........(.....)))))))).)))))...))).... ( -38.00) >consensus GGUCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAAAAUUUCACCGAGUCUCUCGUUGAGACAGUGCCCAAAUCAUUACGCCUUUCGUGCGGGUCGGAACUUACCCGAC (((..(((((.((((.(((((((((((((........)))))................(((((.....)))))..)))))..........(.....)))))))).)))))...))).... (-38.00 = -38.00 + 0.00)


| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -39.05 |
| Consensus MFE | -39.00 |
| Energy contribution | -39.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.05 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/840-960 GUCGGGUAAGUUCCGACCCGCACGAAAGGCGUAACGAUUUGGGCACUGUCUCAACGAGAGACUCGGUGAAAUCAUAGUACCUGUGAAGAUGCAGGUUACCCGCGACAGGACGGAAAGACC ....(((...(((((.(((((.(....))))...((....((((((((((((.....)))))..))))..........((((((......))))))..))).))...)).)))))..))) ( -39.10) >sy_ha.0 1576954 120 - 2685015/840-960 GUCGGGUAAGUUCCGACCCGCACGAAAGGCGUAACGAUUUGGGCACUGUCUCAACGAGAGACUCGGUGAAAUCAUAGUACCUGUGAAGAUGCAGGUUACCCGCGACAGGACGGAAAGACC ....(((...(((((.(((((.(....))))...((....((((((((((((.....)))))..))))..........((((((......))))))..))).))...)).)))))..))) ( -39.10) >sy_sa.0 1551999 120 - 2516575/840-960 GUCGGGUAAGUUCCGACCCGCACGAAAGGCGUAACGAUUUGGGCACUGUCUCAACGAGAGACUCGGUGAAAUCAUAGUACCUGUGAAGAUGCAGGUUACCCGCGACAGGACGGAAAGACC ....(((...(((((.(((((.(....))))...((....((((((((((((.....)))))..))))..........((((((......))))))..))).))...)).)))))..))) ( -39.10) >sp_ag.0 2133292 120 - 2160267/840-960 GUCGGGUAAGUUCCGACCCGCACGAAAGGCGUAAUGAUUUGGGCACUGUCUCAACGAGAGACUCGGUGAAAUUUUAGUACCUGUGAAGAUGCAGGUUACCCGCGACAGGACGGAAAGACC ....(((...(((((.(((((.(....)))).........((((((((((((.....)))))..))))..........((((((......))))))..)))......)).)))))..))) ( -39.00) >sp_pn.0 1692470 120 + 2038615/840-960 GUCGGGUAAGUUCCGACCCGCACGAAAGGCGUAAUGAUUUGGGCACUGUCUCAACGAGAGACUCGGUGAAAUUUUAGUACCUGUGAAGAUGCAGGUUACCCGCGACAGGACGGAAAGACC ....(((...(((((.(((((.(....)))).........((((((((((((.....)))))..))))..........((((((......))))))..)))......)).)))))..))) ( -39.00) >sp_mu.0 1873490 120 + 2030921/840-960 GUCGGGUAAGUUCCGACCCGCACGAAAGGCGUAAUGAUUUGGGCACUGUCUCAACGAGAGACUCGGUGAAAUUUUAGUACCUGUGAAGAUGCAGGUUACCCGCGACAGGACGGAAAGACC ....(((...(((((.(((((.(....)))).........((((((((((((.....)))))..))))..........((((((......))))))..)))......)).)))))..))) ( -39.00) >consensus GUCGGGUAAGUUCCGACCCGCACGAAAGGCGUAACGAUUUGGGCACUGUCUCAACGAGAGACUCGGUGAAAUCAUAGUACCUGUGAAGAUGCAGGUUACCCGCGACAGGACGGAAAGACC ....(((...(((((.(((((.(....)))).........((((((((((((.....)))))..))))..........((((((......))))))..)))......)).)))))..))) (-39.00 = -39.00 + 0.00) # Strand winner: reverse (0.91)

| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.50 |
| Mean single sequence MFE | -35.90 |
| Consensus MFE | -34.50 |
| Energy contribution | -34.25 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -0.73 |
| Structure conservation index | 0.96 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/880-1000 CGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGUUCCGACCCGCACGAAAGGCGUAACGAUUUGGGCACUGUCUCAACGAGAGACUCGGUGAAAUCAUAGUAC .((((((.......))))))..((((..........))))((((((.....)))))).(((.(....))))....(((((.(.(...(((((.....)))))..).).)))))....... ( -35.30) >sy_ha.0 1576954 120 - 2685015/880-1000 CGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGUUCCGACCCGCACGAAAGGCGUAACGAUUUGGGCACUGUCUCAACGAGAGACUCGGUGAAAUCAUAGUAC .((((((.......))))))..((((..........))))((((((.....)))))).(((.(....))))....(((((.(.(...(((((.....)))))..).).)))))....... ( -35.30) >sy_sa.0 1551999 120 - 2516575/880-1000 CGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGUUCCGACCCGCACGAAAGGCGUAACGAUUUGGGCACUGUCUCAACGAGAGACUCGGUGAAAUCAUAGUAC .((((((.......))))))..((((..........))))((((((.....)))))).(((.(....))))....(((((.(.(...(((((.....)))))..).).)))))....... ( -35.30) >sp_ag.0 2133292 120 - 2160267/880-1000 CGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGUUCCGACCCGCACGAAAGGCGUAAUGAUUUGGGCACUGUCUCAACGAGAGACUCGGUGAAAUUUUAGUAC .((((((.......))))))(((((((........(((..(((((((........))))((.(....))).....)))..)))(((((((((.....)))))..))))..)))))))... ( -36.50) >sp_pn.0 1692470 120 + 2038615/880-1000 CGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGUUCCGACCCGCACGAAAGGCGUAAUGAUUUGGGCACUGUCUCAACGAGAGACUCGGUGAAAUUUUAGUAC .((((((.......))))))(((((((........(((..(((((((........))))((.(....))).....)))..)))(((((((((.....)))))..))))..)))))))... ( -36.50) >sp_mu.0 1873490 120 + 2030921/880-1000 CGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGUUCCGACCCGCACGAAAGGCGUAAUGAUUUGGGCACUGUCUCAACGAGAGACUCGGUGAAAUUUUAGUAC .((((((.......))))))(((((((........(((..(((((((........))))((.(....))).....)))..)))(((((((((.....)))))..))))..)))))))... ( -36.50) >consensus CGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGUUCCGACCCGCACGAAAGGCGUAACGAUUUGGGCACUGUCUCAACGAGAGACUCGGUGAAAUCAUAGUAC .((((((.......))))))(((.(((........(((..(((((((........))))((.(....))).....)))..)))(((((((((.....)))))..))))..))).)))... (-34.50 = -34.25 + -0.25) # Strand winner: reverse (0.93)

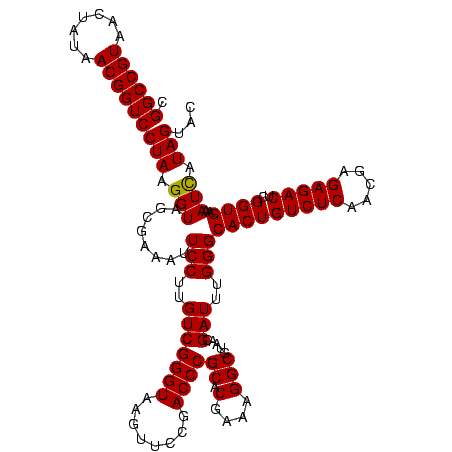
| Location | 1,977,118 – 1,977,236 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -40.48 |
| Consensus MFE | -38.72 |
| Energy contribution | -37.65 |
| Covariance contribution | -1.07 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 118 + 2809422/960-1080 UGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGGUUAG-CUUCUG-CGAAGCUACGAAUCGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUU .(..(((((((...((((((..((((..((...((((((.(-(....)-)..))))))...))..))))))))))...)))((((((.......))))))...))))..).......... ( -41.10) >sy_ha.0 1576954 118 - 2685015/960-1080 UGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGGUUAG-CUUCUG-CGAAGCUACGAAUCGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUU .(..(((((((...((((((..((((..((...((((((.(-(....)-)..))))))...))..))))))))))...)))((((((.......))))))...))))..).......... ( -41.10) >sy_sa.0 1551999 118 - 2516575/960-1080 UGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGGUUAG-CUUCUG-CGAAGCUACGAAUCGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUU .(..(((((((...((((((..((((..((...((((((.(-(....)-)..))))))...))..))))))))))...)))((((((.......))))))...))))..).......... ( -41.10) >sp_ag.0 2133292 118 - 2160267/960-1080 UGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGCUUAG-CGUAAG-CGAAGGUAUGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUU .(..(((((((...((((((..((((..((...((((((.(-(....)-)..))))))...))..))))))))))...)))((((((.......))))))...))))..).......... ( -40.00) >sp_pn.0 1692470 118 + 2038615/960-1080 UGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGCUUAG-CGUAAG-CGAAGGUAUGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUU .(..(((((((...((((((..((((..((...((((((.(-(....)-)..))))))...))..))))))))))...)))((((((.......))))))...))))..).......... ( -40.00) >sp_mu.0 1873490 120 + 2030921/960-1080 UGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGCGCUUAGACGUUUGUCGAAGGUGUGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUU .(..(((((((...((((((..((((..((...((((((.(((....)))..))))))...))..))))))))))...)))((((((.......))))))...))))..).......... ( -39.60) >consensus UGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGCUUAG_CGUCUG_CGAAGCUACGAAUCGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUU .(..(((((((...((((((..((((..((...((((((.(((....)))..))))))...))..))))))))))...)))((((((.......))))))...))))..).......... (-38.72 = -37.65 + -1.07) # Strand winner: reverse (1.00)
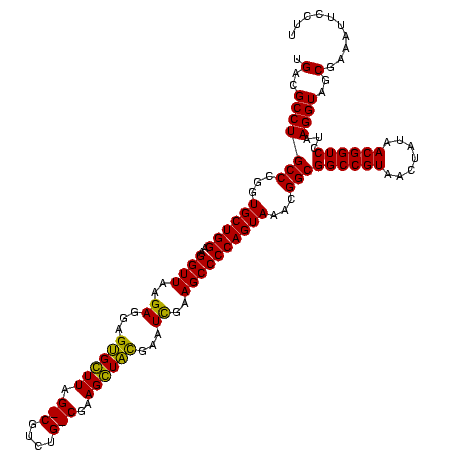
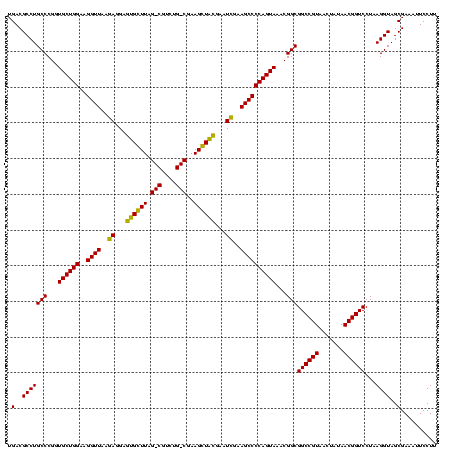
| Location | 1,977,118 – 1,977,236 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.70 |
| Mean single sequence MFE | -39.70 |
| Consensus MFE | -37.28 |
| Energy contribution | -35.22 |
| Covariance contribution | -2.07 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 118 + 2809422/1000-1120 ACACAGGUCUCUGCUAAACCGUAAGGUGAUGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGGUUAG-CUUCUG-CGAAGCUACGAAUCGAAGCCCCAGUAAACGG ...(((((.((.(((..(((....)))..........))).)).))))).(((.((((((..((((..((...((((((.(-(....)-)..))))))...))..))))))))))..))) ( -41.30) >sy_ha.0 1576954 118 - 2685015/1000-1120 ACACAGGUCUCUGCUAAACCGUAAGGUGAUGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGGUUAG-CUUCUG-CGAAGCUACGAAUCGAAGCCCCAGUAAACGG ...(((((.((.(((..(((....)))..........))).)).))))).(((.((((((..((((..((...((((((.(-(....)-)..))))))...))..))))))))))..))) ( -41.30) >sy_sa.0 1551999 118 - 2516575/1000-1120 ACACAGGUCUCUGCUAAACCGUAAGGUGAUGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGGUUAG-CUUCUG-CGAAGCUACGAAUCGAAGCCCCAGUAAACGG ...(((((.((.(((..(((....)))..........))).)).))))).(((.((((((..((((..((...((((((.(-(....)-)..))))))...))..))))))))))..))) ( -41.30) >sp_ag.0 2133292 118 - 2160267/1000-1120 ACACAGCUCUCUGCUAAAUCGUAAGAUGAUGUAUAGGGGGUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGCUUAG-CGUAAG-CGAAGGUAUGAAUUGAAGCCCCAGUAAACGG .(((..((((.(((...(((....)))...))).)))).)))........(((.((((((..((((..((...((((((.(-(....)-)..))))))...))..))))))))))..))) ( -38.10) >sp_pn.0 1692470 118 + 2038615/1000-1120 ACACAGCUCUCUGCUAAAUCGUAAGAUGAUGUAUAGGGGGUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGCUUAG-CGUAAG-CGAAGGUAUGAAUUGAAGCCCCAGUAAACGG .(((..((((.(((...(((....)))...))).)))).)))........(((.((((((..((((..((...((((((.(-(....)-)..))))))...))..))))))))))..))) ( -38.10) >sp_mu.0 1873490 120 + 2030921/1000-1120 ACACAGCUCUCUGCGAAAUCGUAAGAUGAAGUAUAGGGGGUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGCGCUUAGACGUUUGUCGAAGGUGUGAAUUGAAGCCCCAGUAAACGG .(((..((((.(((...(((....)))...))).)))).)))........(((.((((((..((((..((...((((((.(((....)))..))))))...))..))))))))))..))) ( -38.10) >consensus ACACAGCUCUCUGCUAAACCGUAAGAUGAUGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGCUUAG_CGUCUG_CGAAGCUACGAAUCGAAGCCCCAGUAAACGG .....(((((((((...(((....)))...))..))))))).........(((.((((((..((((..((...((((((.(((....)))..))))))...))..))))))))))..))) (-37.28 = -35.22 + -2.07) # Strand winner: reverse (0.99)

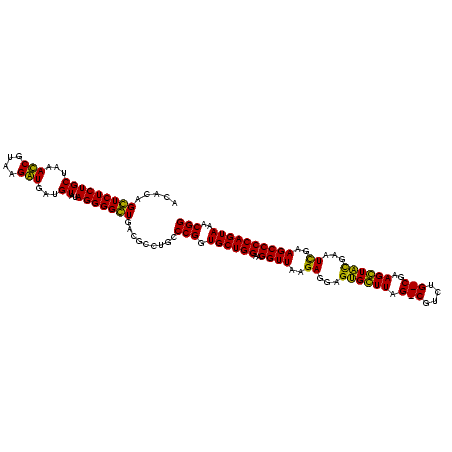
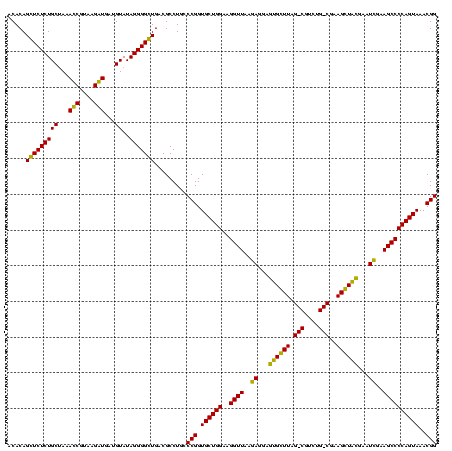
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.28 |
| Mean single sequence MFE | -35.52 |
| Consensus MFE | -24.73 |
| Energy contribution | -22.70 |
| Covariance contribution | -2.03 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/1440-1560 UAGUCGGGUCCUAAGCUGAGGCCGACAGGCGUAGGCGAUGGAUAACAGGUUGAUAUUCCUGUACCACCUAUAAUCGUUUUAAUCGAUGGGGGGACGCAGUAGGAUAGGCGAAGCGUGCGA ..(((((((((...(((.(.(((....))).).)))...)))).(((((........))))).)).(((((.((((.......))))(.(....).).)))))...)))........... ( -38.20) >sy_ha.0 1576954 120 - 2685015/1440-1560 UAGUCGGGUCCUAAGCUGAGGCCGACAGGCGUAGGCGAUGGAUAACAGGUUGAUAUUCCUGUACCACCUAGCAUCGUUUUAAUCGAUGGGGGGACGCAGUAGGAUAGGCGAAGCGUACGA ..(((..((((((..(((..(((....))).)))(((.(((...(((((........))))).)))(((..(((((.......)))))..))).)))..)))))).)))........... ( -39.70) >sy_sa.0 1551999 120 - 2516575/1440-1560 UAGUCGGGUCCUAAGCUGAGGCCGAUAGGCGUAGGCGAUGGAUAACAGGUUGAUAUUCCUGUACCACCAUUAUUCGUUUUAAGCGAUGGGGGGACGCAGUAGGAUAGGCGAAGCGUACGA ..(((((..(((......))))))))...((((.(((((((...(((((........)))))....))))).(((((((.......((.(....).)).......))))))))).)))). ( -38.24) >sp_ag.0 2133292 113 - 2160267/1440-1560 UAGUCGGGACCUAAGGAGAGACCGAAAGGUGUAUCCGAUGGACAACAGGUUGAUAUUCCUGUACUA-------GAGUAUAUAGUGAUGGAGGGACGCAGUAGGCUAACUAAACCAGACGA ..(((.((.((...(((.(.(((....))).).)))...))......(((((...((((..(((((-------.......)))))..))))....((.....)))))))...)).))).. ( -30.70) >sp_pn.0 1692470 113 + 2038615/1440-1560 UAGUCGGGACCUAAGGAGAGACCGAAAGGUGUAUCCGAUGGACAACAGGUUGAUAUUCCUGUACUA-------GAGUAUGUAGUGAUGGAGGGACGCAGUAGGCUAACUAAAGCAGACGA ..(((.(..(((..(((.(.(((....))).).)))......(((((((........)))))((((-------.......))))..)).)))..)((..(((.....)))..)).))).. ( -32.00) >sp_mu.0 1873490 113 + 2030921/1440-1560 UAGUCGGGACCUAAGGAGAGACCGAUAGGUGUAUCCGAUGGGCAACAGGUUGAUAUUCCUGUACUA-------GAGUAUUGAGUGAAGGAGGGACGCAGCAGGCUAACUAGAGCGUGCGA ..(((..(((((..(((.(.(((....))).).)))....(....))))))....(((((.((((.-------........)))).))))).)))...(((.(((......))).))).. ( -34.30) >consensus UAGUCGGGACCUAAGCAGAGACCGAAAGGCGUAGCCGAUGGACAACAGGUUGAUAUUCCUGUACCA_______GAGUAUUAAGCGAUGGAGGGACGCAGUAGGAUAACCAAAGCGUACGA ......(..(((..(((.(.(((....))).).)))..(((...(((((........))))).))).......................)))..)...(((.(..........).))).. (-24.73 = -22.70 + -2.03) # Strand winner: reverse (1.00)

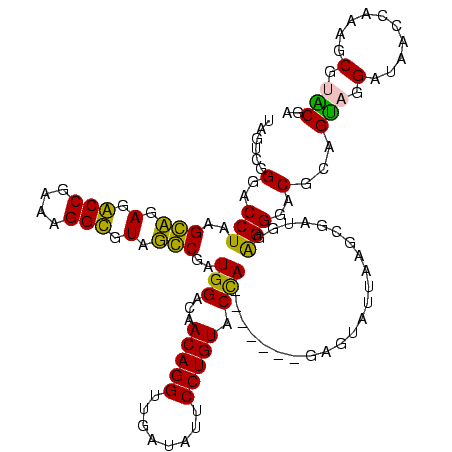
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.06 |
| Mean single sequence MFE | -32.42 |
| Consensus MFE | -27.48 |
| Energy contribution | -25.98 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/1480-1600 AAAACGAUUAUAGGUGGUACAGGAAUAUCAACCUGUUAUCCAUCGCCUACGCCUGUCGGCCUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAA .....((((((((((((((((((........))))).....)))))))).....((((((((......)))..)))))....((((((.(((......)))))))))......))))).. ( -36.20) >sy_ha.0 1576954 120 - 2685015/1480-1600 AAAACGAUGCUAGGUGGUACAGGAAUAUCAACCUGUUAUCCAUCGCCUACGCCUGUCGGCCUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAA .....((((((((((((((((((........))))).....)))))))).))..)))(((((......))).)).((((((.((((((.(((......))))))))).....)))))).. ( -37.20) >sy_sa.0 1551999 120 - 2516575/1480-1600 AAAACGAAUAAUGGUGGUACAGGAAUAUCAACCUGUUAUCCAUCGCCUACGCCUAUCGGCCUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAA ............(((((.(((((........)))))...)))))((....(((....)))....)).........((((((.((((((.(((......))))))))).....)))))).. ( -33.60) >sp_ag.0 2133292 113 - 2160267/1480-1600 UAUACUC-------UAGUACAGGAAUAUCAACCUGUUGUCCAUCGGAUACACCUUUCGGUCUCUCCUUAGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAACCUUAGUCUU .......-------....(((((........)))))(((((...))))).((((...((.....))..))))...((((((.((((((.(((......))))))))).....)))))).. ( -28.90) >sp_pn.0 1692470 113 + 2038615/1480-1600 CAUACUC-------UAGUACAGGAAUAUCAACCUGUUGUCCAUCGGAUACACCUUUCGGUCUCUCCUUAGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAACCUUAGUCUU .......-------....(((((........)))))(((((...))))).((((...((.....))..))))...((((((.((((((.(((......))))))))).....)))))).. ( -28.90) >sp_mu.0 1873490 113 + 2030921/1480-1600 AAUACUC-------UAGUACAGGAAUAUCAACCUGUUGCCCAUCGGAUACACCUAUCGGUCUCUCCUUAGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAUCCUUAGUCUU .......-------..(((((((........)))).))).....((....(((((..((.....)).))))).))((((((.((((((.(((......))))))))).....)))))).. ( -29.70) >consensus AAAACGA_______UAGUACAGGAAUAUCAACCUGUUAUCCAUCGCAUACACCUAUCGGCCUCACCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCCCUGGAAACCUUAGUCAA ..................(((((........)))))...((...((....(((....)))....))...))....((((((.((((((.(((......))))))))).....)))))).. (-27.48 = -25.98 + -1.50)
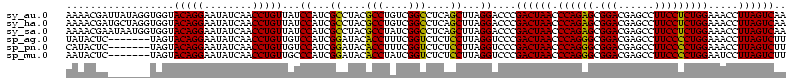
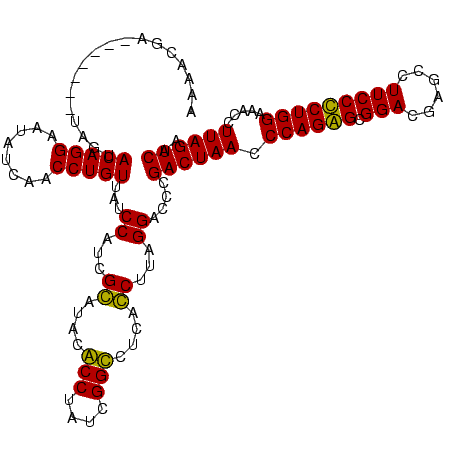
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.06 |
| Mean single sequence MFE | -39.20 |
| Consensus MFE | -39.58 |
| Energy contribution | -37.33 |
| Covariance contribution | -2.25 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.42 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/1480-1600 UUGACUAAGGUUUCCAGAGGAAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGACAGGCGUAGGCGAUGGAUAACAGGUUGAUAUUCCUGUACCACCUAUAAUCGUUUU ..((((((....(((((((...((.....)).)))))))))))))((((((...(((.(.(((....))).).)))...)))).(((((........))))).))............... ( -40.30) >sy_ha.0 1576954 120 - 2685015/1480-1600 UUGACUAAGGUUUCCAGAGGAAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGACAGGCGUAGGCGAUGGAUAACAGGUUGAUAUUCCUGUACCACCUAGCAUCGUUUU ..((((((....(((((((...((.....)).)))))))))))))((((((...(((.(.(((....))).).)))...)))).(((((........))))).))............... ( -40.30) >sy_sa.0 1551999 120 - 2516575/1480-1600 UUGACUAAGGUUUCCAGAGGAAGGCUCGUCCGCUCUGGGUUAGUCGGGUCCUAAGCUGAGGCCGAUAGGCGUAGGCGAUGGAUAACAGGUUGAUAUUCCUGUACCACCAUUAUUCGUUUU ..((((((....(((((((...((.....)).)))))))))))))((((((...(((.(.(((....))).).)))...)))).(((((........))))).))............... ( -40.30) >sp_ag.0 2133292 113 - 2160267/1480-1600 AAGACUAAGGUUUCCAGGGGAAGGCUCGUCCGCCCUGGGUUAGUCGGGACCUAAGGAGAGACCGAAAGGUGUAUCCGAUGGACAACAGGUUGAUAUUCCUGUACUA-------GAGUAUA ..((((((....(((((((...((.....)).)))))))))))))....((...(((.(.(((....))).).)))...))...(((((........)))))....-------....... ( -38.70) >sp_pn.0 1692470 113 + 2038615/1480-1600 AAGACUAAGGUUUCCAGGGGAAGGCUCGUCCGCCCUGGGUUAGUCGGGACCUAAGGAGAGACCGAAAGGUGUAUCCGAUGGACAACAGGUUGAUAUUCCUGUACUA-------GAGUAUG ..((((((....(((((((...((.....)).)))))))))))))....((...(((.(.(((....))).).)))...))...(((((........)))))....-------....... ( -38.70) >sp_mu.0 1873490 113 + 2030921/1480-1600 AAGACUAAGGAUUCCAGGGGAAGGCUCGUCCGCCCUGGGUUAGUCGGGACCUAAGGAGAGACCGAUAGGUGUAUCCGAUGGGCAACAGGUUGAUAUUCCUGUACUA-------GAGUAUU ..((((((....(((((((...((.....)).)))))))))))))..(.((((.(((.(.(((....))).).)))..))))).(((((........)))))....-------....... ( -36.90) >consensus AAGACUAAGGUUUCCAGAGGAAGGCUCGUCCGCCCUGGGUUAGUCGGGACCUAAGCAGAGACCGAAAGGCGUAGCCGAUGGACAACAGGUUGAUAUUCCUGUACCA_______GAGUAUU ..((((((....(((((((...((.....)).)))))))))))))((..((...(((.(.(((....))).).)))...))...(((((........))))).))............... (-39.58 = -37.33 + -2.25) # Strand winner: reverse (1.00)
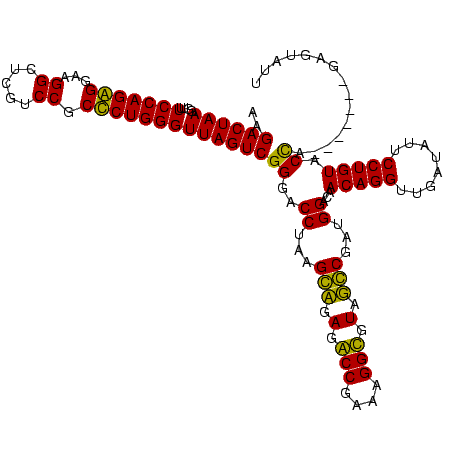
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.06 |
| Mean single sequence MFE | -36.35 |
| Consensus MFE | -35.60 |
| Energy contribution | -34.10 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/1520-1640 CAUCGCCUACGCCUGUCGGCCUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACAC ....((...((((....(((((......))).)).((((((.((((((.(((......))))))))).....))))))....))))((((((.......))))))....))......... ( -36.60) >sy_ha.0 1576954 120 - 2685015/1520-1640 CAUCGCCUACGCCUGUCGGCCUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACAC ....((...((((....(((((......))).)).((((((.((((((.(((......))))))))).....))))))....))))((((((.......))))))....))......... ( -36.60) >sy_sa.0 1551999 120 - 2516575/1520-1640 CAUCGCCUACGCCUAUCGGCCUCAGCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCUCUGGAAACCUUAGUCAAUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACAC ....((...((((....(((((......))).)).((((((.((((((.(((......))))))))).....))))))....))))((((((.......))))))....))......... ( -36.60) >sp_ag.0 2133292 120 - 2160267/1520-1640 CAUCGGAUACACCUUUCGGUCUCUCCUUAGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAACCUUAGUCUUACGGUGGACAGGAUUCUCACCUGUCUUGCGCUACUCAUAC (((((.....((((...((.....))..))))...((((((.((((((.(((......))))))))).....))))))...)))))((((((.......))))))............... ( -35.70) >sp_pn.0 1692470 120 + 2038615/1520-1640 CAUCGGAUACACCUUUCGGUCUCUCCUUAGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAACCUUAGUCUUACGGUGGACAGGAUUCUCACCUGUCUUUCGCUACUCAUAC ...((((..((((....((..(((....)))..))((((((.((((((.(((......))))))))).....))))))....))))((((((.......)))))).)))).......... ( -35.80) >sp_mu.0 1873490 120 + 2030921/1520-1640 CAUCGGAUACACCUAUCGGUCUCUCCUUAGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAUCCUUAGUCUUACGGUGGACAGGAUUCUCACCUGUCUUACGCUACUCAUAC (((((.....(((((..((.....)).)))))...((((((.((((((.(((......))))))))).....))))))...)))))((((((.......))))))............... ( -36.80) >consensus CAUCGCAUACACCUAUCGGCCUCACCUUAGGACCCGACUAACCCAGAGCGGACGAGCCUUCCCCUGGAAACCUUAGUCAAACGGUGGACAGGAUUCUCACCCGUCUUUCGCUACUCACAC .........((((....(((((......))).)).((((((.((((((.(((......))))))))).....))))))....))))((((((.......))))))............... (-35.60 = -34.10 + -1.50)

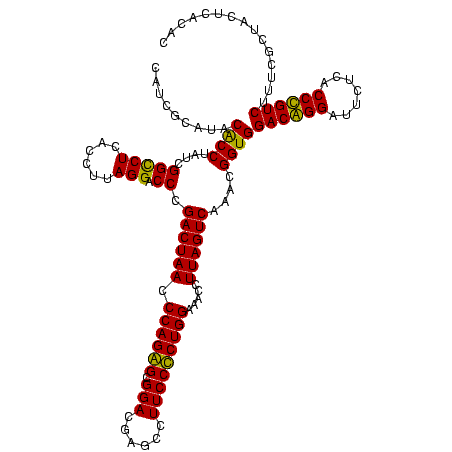
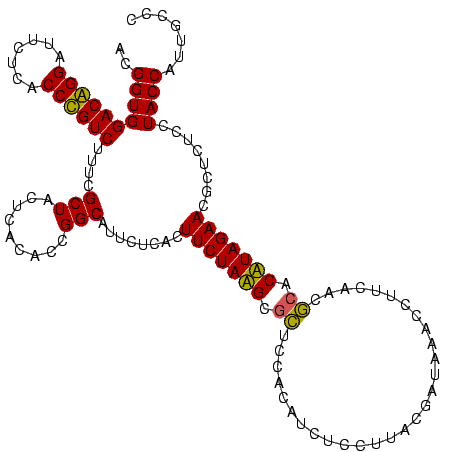
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.98 |
| Mean single sequence MFE | -25.00 |
| Consensus MFE | -21.18 |
| Energy contribution | -19.90 |
| Covariance contribution | -1.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.566704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/1600-1720 UCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUACGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCC ((((((((((((.......))))))....(.....).)))))).........(((((((.((....((((((.....)).)))).......)).)))))))................... ( -25.60) >sy_ha.0 1576954 120 - 2685015/1600-1720 UCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUGCAAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCC ((((((((((((.......))))))....(.....).)))))).........(((((((.((....((((..........)))).......)).)))))))................... ( -24.50) >sy_sa.0 1551999 120 - 2516575/1600-1720 UCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUGCGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCC ((((((((((((.......))))))....(.....).)))))).........(((((((.((....((((((.....)).)))).......)).)))))))................... ( -25.60) >sp_ag.0 2133292 120 - 2160267/1600-1720 ACGGUGGACAGGAUUCUCACCUGUCUUGCGCUACUCAUACCGGCAUUCUCACUUCUAUGCGUUCCAGCGCUCCUCACGGUACACCUUCUUCACACAUAGAACGCUCUCCUACCAUGACAC ..((((((((((.......))))))..(((((((.((((..((........))..)))).))...)))))...........))))((((........))))................... ( -25.60) >sp_pn.0 1692470 118 + 2038615/1600-1720 ACGGUGGACAGGAUUCUCACCUGUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAUGCGUUCCAGCACUCCUCACGGUAUACCUUCAUCACACAUAGAACGCUCUCCUACCAUACC-- ..((((((((((.......))))))....((.....((((((((((..........))))((....))........)))))).......((.......))..)).....)))).....-- ( -23.30) >sp_mu.0 1873490 118 + 2030921/1600-1720 ACGGUGGACAGGAUUCUCACCUGUCUUACGCUACUCAUACCGGCAUUCUCACUUCUAUGCACUCCAGCGCUCCUUGCGGUACACCUUCUGCGCACAUAGAACGCUCUCCUACCAUCCC-- ..((((((((((.......))))))............((((.((((..........))))........((.....))))))))))....(((.........)))..............-- ( -25.40) >consensus ACGGUGGACAGGAUUCUCACCCGUCUUUCGCUACUCACACCGGCAUUCUCACUUCUAAGCGCUCCACAUCUCCUUACGAUAAACCUUCAACGCACAUAGAACGCUCUCCUACCAUUGCCC ..((((((((((.......))))))....(((.........)))........(((((((.((.............................)).)))))))........))))....... (-21.18 = -19.90 + -1.28)
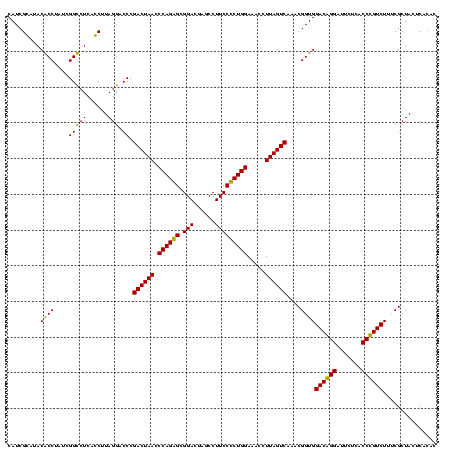
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.98 |
| Mean single sequence MFE | -41.30 |
| Consensus MFE | -40.51 |
| Energy contribution | -36.60 |
| Covariance contribution | -3.91 |
| Combinations/Pair | 1.41 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.18 |
| SVM RNA-class probability | 0.998657 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/1600-1720 GGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGUAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGA (((((.(((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))).)))...........((((((.......))))))..)).. ( -41.90) >sy_ha.0 1576954 120 - 2685015/1600-1720 GGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUUGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGA (((((.(((((.....((.(((((((.((......(((((..........)))))...)).))))))))).....))))).)))...........((((((.......))))))..)).. ( -37.50) >sy_sa.0 1551999 120 - 2516575/1600-1720 GGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCGGUGUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGA (((((.(((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))).)))...........((((((.......))))))..)).. ( -42.90) >sp_ag.0 2133292 120 - 2160267/1600-1720 GUGUCAUGGUAGGAGAGCGUUCUAUGUGUGAAGAAGGUGUACCGUGAGGAGCGCUGGAACGCAUAGAAGUGAGAAUGCCGGUAUGAGUAGCGCAAGACAGGUGAGAAUCCUGUCCACCGU ...((((((((.....((.((((((((((......(((((.((....)).)))))...)))))))))))).....))))...))))...(((...((((((.......))))))...))) ( -41.90) >sp_pn.0 1692470 118 + 2038615/1600-1720 --GGUAUGGUAGGAGAGCGUUCUAUGUGUGAUGAAGGUAUACCGUGAGGAGUGCUGGAACGCAUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGACAGGUGAGAAUCCUGUCCACCGU --(((.(((((.....((.((((((((((......(((((.((....)).)))))...)))))))))))).....)))))...............((((((.......)))))).))).. ( -41.40) >sp_mu.0 1873490 118 + 2030921/1600-1720 --GGGAUGGUAGGAGAGCGUUCUAUGUGCGCAGAAGGUGUACCGCAAGGAGCGCUGGAGUGCAUAGAAGUGAGAAUGCCGGUAUGAGUAGCGUAAGACAGGUGAGAAUCCUGUCCACCGU --((..(((((.....((.(((((((..(.(....(((((.((....)).))))).).)..))))))))).....)))))...............((((((.......))))))..)).. ( -42.20) >consensus GGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGACCGCAAGGACAGCUGGAGCGCAUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGACAGGUGAGAAUCCCGUCCACCGA ..(((.(((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))).)))......((...((((((.......))))))...)). (-40.51 = -36.60 + -3.91) # Strand winner: reverse (1.00)
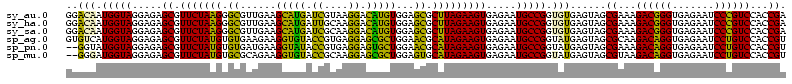


| Location | 1,977,118 – 1,977,236 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -23.00 |
| Consensus MFE | -20.77 |
| Energy contribution | -18.72 |
| Covariance contribution | -2.05 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 118 + 2809422/1640-1760 CGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUACGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCCA-AAGGACAAUCCACAGCUUCGGUAAUAUGUUU-AGCCCC .(((........(((((((.((....((((((.....)).)))).......)).)))))))........(((((((((((.-..)))))))..........))))........-.))).. ( -23.20) >sy_ha.0 1576954 118 - 2685015/1640-1760 CGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUGCAAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCCU-ACGGACAAUCCACAGCUUCGGUAAUAUGUUU-AGCCCC .(((........(((((((.((....((((..........)))).......)).)))))))........(((((((((((.-..)))))))..........))))........-.))).. ( -22.10) >sy_sa.0 1551999 119 - 2516575/1640-1760 CGGCAUUCUCACUUCUAAGCGCUCCACAUGUCCUUGCGAUCAUGCUUCAACGCCCUUAGAACGCUCUCCUACCAUUGUCCUUACGGACAAUCCACAGCUUCGGUAAUAUGUUU-AGCCCC .(((........(((((((.((....((((((.....)).)))).......)).)))))))........(((((((((((....)))))))..........))))........-.))).. ( -24.70) >sp_ag.0 2133292 120 - 2160267/1640-1760 CGGCAUUCUCACUUCUAUGCGUUCCAGCGCUCCUCACGGUACACCUUCUUCACACAUAGAACGCUCUCCUACCAUGACACUUUUGUGUCAUCCACAGCUUCGGUAAUAUGUUUUAGCCCC .(((..............((((....)))).......((....))...........(((((((..........(((((((....))))))).....((....))....)))))))))).. ( -24.90) >sp_pn.0 1692470 118 + 2038615/1640-1760 CGGCAUUCUCACUUCUAUGCGUUCCAGCACUCCUCACGGUAUACCUUCAUCACACAUAGAACGCUCUCCUACCAUACC--UAUAAAGGUAUCCACAGCUUCGGUAAAUUGUUUUAGCCCC .(((..............((((((.............((....)).............)))))).....(((((((((--(....))))))..........))))..........))).. ( -20.37) >sp_mu.0 1873490 116 + 2030921/1640-1760 CGGCAUUCUCACUUCUAUGCACUCCAGCGCUCCUUGCGGUACACCUUCUGCGCACAUAGAACGCUCUCCUACCAUCCC----CUAAGGGAUCCACAGCUUCGGUAAAUUGUUUCAGCCCC .(((........(((((((........(((.....)))(..((.....))..).)))))))........(((((((((----....)))))..........))))..........))).. ( -22.70) >consensus CGGCAUUCUCACUUCUAAGCGCUCCACAUCUCCUUACGAUAAACCUUCAACGCACAUAGAACGCUCUCCUACCAUUGCCCU_AAGGACAAUCCACAGCUUCGGUAAUAUGUUU_AGCCCC .(((........(((((((.((.............................)).)))))))........(((((((((((....)))))))..........))))..........))).. (-20.77 = -18.72 + -2.05)

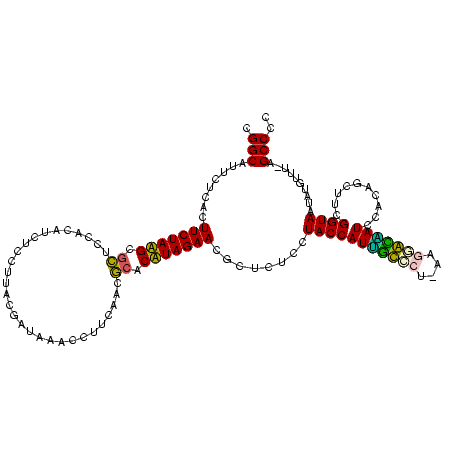
| Location | 1,977,118 – 1,977,236 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -38.27 |
| Consensus MFE | -40.07 |
| Energy contribution | -35.21 |
| Covariance contribution | -4.86 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.70 |
| Structure conservation index | 1.05 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994250 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 118 + 2809422/1640-1760 GGGGCU-AAACAUAUUACCGAAGCUGUGGAUUGUCCUU-UGGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGUAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCG ..((((-..............))))....(((((((..-.)))))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....)))). ( -35.54) >sy_ha.0 1576954 118 - 2685015/1640-1760 GGGGCU-AAACAUAUUACCGAAGCUGUGGAUUGUCCGU-AGGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUUGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCG ..(((.-...(((.(((((....(....)(((((((..-.))))))))))))..((((((((((.....(((...(((....)))...)))...))))))))))....))).....))). ( -32.30) >sy_sa.0 1551999 119 - 2516575/1640-1760 GGGGCU-AAACAUAUUACCGAAGCUGUGGAUUGUCCGUAAGGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGAUCGCAAGGACAUGUGGAGCGCUUAGAAGUGAGAAUGCCG ..((((-..............))))....(((((((....)))))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....)))). ( -40.84) >sp_ag.0 2133292 120 - 2160267/1640-1760 GGGGCUAAAACAUAUUACCGAAGCUGUGGAUGACACAAAAGUGUCAUGGUAGGAGAGCGUUCUAUGUGUGAAGAAGGUGUACCGUGAGGAGCGCUGGAACGCAUAGAAGUGAGAAUGCCG ..((((...............))))....(((((((....)))))))((((.....((.((((((((((......(((((.((....)).)))))...)))))))))))).....)))). ( -41.16) >sp_pn.0 1692470 118 + 2038615/1640-1760 GGGGCUAAAACAAUUUACCGAAGCUGUGGAUACCUUUAUA--GGUAUGGUAGGAGAGCGUUCUAUGUGUGAUGAAGGUAUACCGUGAGGAGUGCUGGAACGCAUAGAAGUGAGAAUGCCG ..(((........(((((((.(.(((((((....))))))--).).)))))))...((.((((((((((......(((((.((....)).)))))...))))))))))))......))). ( -39.20) >sp_mu.0 1873490 116 + 2030921/1640-1760 GGGGCUGAAACAAUUUACCGAAGCUGUGGAUCCCUUAG----GGGAUGGUAGGAGAGCGUUCUAUGUGCGCAGAAGGUGUACCGCAAGGAGCGCUGGAGUGCAUAGAAGUGAGAAUGCCG ..((((...............))))....(((((....----)))))((((.....((.(((((((..(.(....(((((.((....)).))))).).)..))))))))).....)))). ( -40.56) >consensus GGGGCU_AAACAUAUUACCGAAGCUGUGGAUUGCCCAU_AGGACAAUGGUAGGAGAGCGUUCUAAGGGCGUUGAAGCAUGACCGCAAGGACAGCUGGAGCGCAUAGAAGUGAGAAUGCCG ..((((...............))))....(((((((....)))))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....)))). (-40.07 = -35.21 + -4.86) # Strand winner: reverse (1.00)

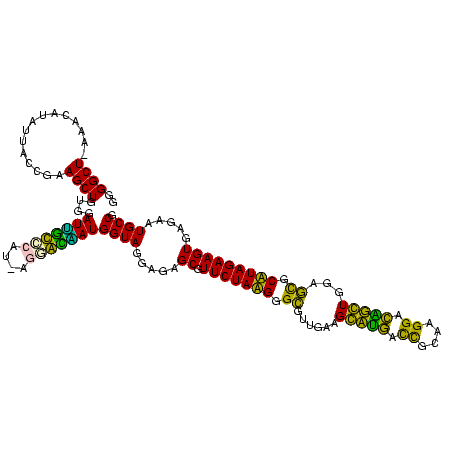

| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -36.03 |
| Consensus MFE | -38.12 |
| Energy contribution | -35.15 |
| Covariance contribution | -2.97 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.02 |
| Structure conservation index | 1.06 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.953723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/2080-2200 CAGGUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAACUGAGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUGGUUCUCUCC ..((((...((((((......))))))....))))...(((.(((((........)))))))).......(((((.(.(((.....(((((........)))))....))).).))))). ( -34.80) >sy_ha.0 1576954 120 - 2685015/2080-2200 CAGGUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAACUGUGGGUAGCGGAGAAAUUCCAAUCGAACUUGGAGAUAGCUGGUUCUCUCC .........((((((......))))))(((((((((..(((.(((((........)))))))).....((((.....)))).....((((((......)))))).....))))))))).. ( -35.60) >sy_sa.0 1551999 120 - 2516575/2080-2200 CAGGUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAGUUGUGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUGGUUCUCUCC .........((((((......))))))((((((((...(((.(((((........))))))))...((((((((((.(.((((...))))...)..))))....)))))))))))))).. ( -35.10) >sp_ag.0 2133292 120 - 2160267/2080-2200 CAGGUUGAAGGUGAGGUAAAACUCACUGGAGGACCGAACCAGGGCACGUUGAAAAGUGCUUGGAUGACUUGUGGGUAGCGGAGAAAUUCCAAACGAACUUGGAGAUAGCUGGUUCUCUCC .........((((((......))))))((((((((...(((((.(((........)))))))).............(((.......((((((......))))))...))))))))))).. ( -36.60) >sp_pn.0 1692470 120 + 2038615/2080-2200 CAGGUUGAAGGUGCGGUAAGACGCACUGGAGGACCGAACCAGGGCACGUUGAAAAGUGCUUGGAUGACUUGUGGGUAGCGGAGAAAUUCCAAACGAACUUGGAGAUAGCUGGUUCUCUCC .........((((((......))))))((((((((...(((((.(((........)))))))).............(((.......((((((......))))))...))))))))))).. ( -37.90) >sp_mu.0 1873490 120 + 2030921/2080-2200 CAGGUUGAAGGUGCGGUAAAACGCACUGGAGGACCGAACCAGGACACGUUGAAAAGUGUUUGGAUGACUUGUGGGUAGCGGAGAAAUUCCAAACGAACUUGGAGAUAGCUGGUUCUCUCC ..((((...((((((......))))))....))))(((((((.....(((...(((((((((((...(((((.....)).)))....)))))))..))))...)))..)))))))..... ( -36.20) >consensus CAGGUUGAAGGUCAGGUAACACUCAAUGGAGGACCGAACCAACUCACGUUGAAAAGUGAGCGGAUGACUUGUGGGUAGCGGAGAAAUUCCAAACGAACUUGGAGAUAGCUGGUUCUCUCC .........((((((......))))))((((((((...(((.(((((........)))))))).............(((.......((((((......))))))...))))))))))).. (-38.12 = -35.15 + -2.97) # Strand winner: reverse (1.00)

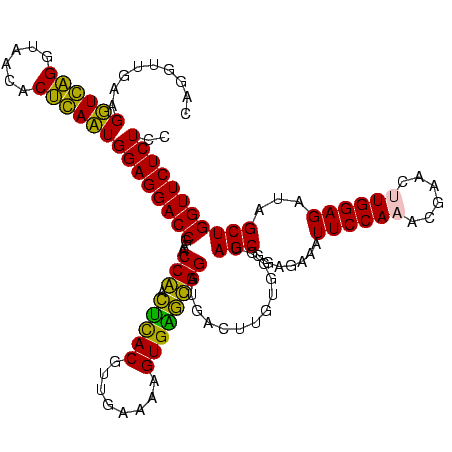
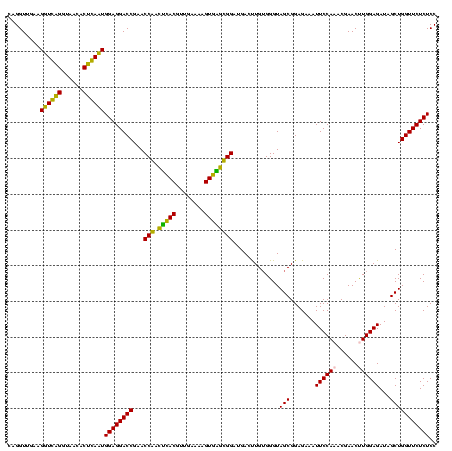
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.67 |
| Mean single sequence MFE | -43.17 |
| Consensus MFE | -44.65 |
| Energy contribution | -40.02 |
| Covariance contribution | -4.63 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.78 |
| Structure conservation index | 1.03 |
| SVM decision value | 3.86 |
| SVM RNA-class probability | 0.999667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/2120-2240 AUUUGGUCGUAGACCCGAAACCAGGUGAUCUACCCUUGGUCAGGUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAACUGAGGGUAGCG (((((((((......)))..))))))...(((((((..((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))..))))))).. ( -44.10) >sy_ha.0 1576954 120 - 2685015/2120-2240 AUUUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGGUCAGGUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAACUGUGGGUAGCG (((((((((......)))..))))))...(((((((((((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))))))))))).. ( -43.80) >sy_sa.0 1551999 120 - 2516575/2120-2240 AUUUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGACCAGGUUGAAGUUCAGGUAACACUGAAUGGAGGACCGAACCGACUUACGUUGAAAAGUGAGCGGAUGAGUUGUGGGUAGCG (((((((((......)))..))))))...(((((((..((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))..))))))).. ( -44.00) >sp_ag.0 2133292 120 - 2160267/2120-2240 AUCAUGUUGUAGACCCGAAACCAUGUGACCUACCCAUGAGCAGGUUGAAGGUGAGGUAAAACUCACUGGAGGACCGAACCAGGGCACGUUGAAAAGUGCUUGGAUGACUUGUGGGUAGCG .((((((.((.........)).)))))).(((((((..((((((((...((((((......))))))....))))...(((((.(((........)))))))).)).))..))))))).. ( -43.60) >sp_pn.0 1692470 120 + 2038615/2120-2240 AUCAUGACGUAGACCCGAAACCAUGUGACCUACCCAUGAGCAGGUUGAAGGUGCGGUAAGACGCACUGGAGGACCGAACCAGGGCACGUUGAAAAGUGCUUGGAUGACUUGUGGGUAGCG ..((((.((......))....))))....(((((((..((((((((...((((((......))))))....))))...(((((.(((........)))))))).)).))..))))))).. ( -43.10) >sp_mu.0 1873490 120 + 2030921/2120-2240 ACGUAGAUGUAGACCCGAAACCAAGUGACCUACCCAUGAGCAGGUUGAAGGUGCGGUAAAACGCACUGGAGGACCGAACCAGGACACGUUGAAAAGUGUUUGGAUGACUUGUGGGUAGCG .............................(((((((..((((((((...((((((......))))))....))))...((((.((((........)))))))).)).))..))))))).. ( -40.40) >consensus AUUUGGUCGUAGACCCGAAACCAGGUGACCUACCCAUGAGCAGGUUGAAGGUCAGGUAACACUCAAUGGAGGACCGAACCAACUCACGUUGAAAAGUGAGCGGAUGACUUGUGGGUAGCG .............................(((((((..((((((((...((((((......))))))....))))...(((.(((((........)))))))).)).))..))))))).. (-44.65 = -40.02 + -4.63) # Strand winner: reverse (1.00)

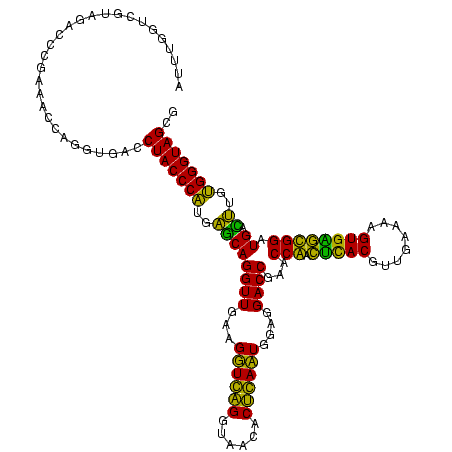
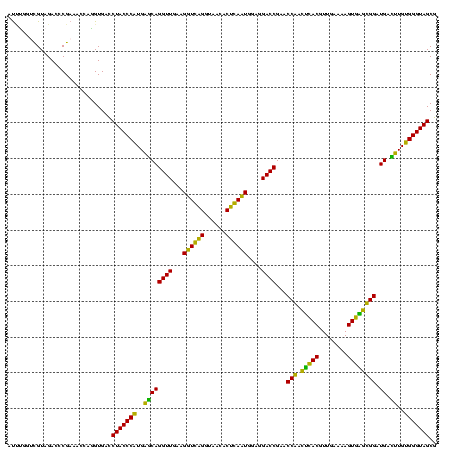
| Location | 1,977,118 – 1,977,237 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.59 |
| Mean single sequence MFE | -37.53 |
| Consensus MFE | -36.04 |
| Energy contribution | -32.30 |
| Covariance contribution | -3.74 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 119 + 2809422/2200-2320 CGGCGAGUUACGAUUUGAUGCAAGGUUAAGCAGUAAAUGUGGAGCCGUAGCGAAAGCGAGUCUGAAUAGGGCG-UUUAGUAUUUGGUCGUAGACCCGAAACCAGGUGAUCUACCCUUGGU (((....(((((((..(((((..((((..(((.....)))..))))...((....))..((((.....)))).-....)))))..)))))))..)))..((((((.(......))))))) ( -37.90) >sy_ha.0 1576954 119 - 2685015/2200-2320 CGGCGAGUUACGAUCUGAUGCAAGGUUAAGCAGGAAAUGUGGAGCCGUAGCGAAAGCGAGUCUGAAUAGGGCG-UUGAGUAUUUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGGU (((....((((((((.(((((..((((..(((.....)))..))))...((....))..((((.....)))).-....))))).))))))))..)))..((((((........)).)))) ( -37.90) >sy_sa.0 1551999 119 - 2516575/2200-2320 CGGCGAGUUACGAUUUGAUGCAAGGUUAAGCAGUGAAUGUGGAGCCGUAGCGAAAGCGAGUCUGAAUAGGGCG-UUGAGUAUUUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGAC (((....(((((((..(((((..((((..(((.....)))..))))...((....))..((((.....)))).-....)))))..)))))))..)))......((((...))))...... ( -33.50) >sp_ag.0 2133292 120 - 2160267/2200-2320 CGGCGAGUUACGAUAUGAUGCGAGGUUAAGUUGAAGAGACGGAGCCGUAGGGAAACCGAGUCUUAAUAGGGCGUCAUAGUAUCAUGUUGUAGACCCGAAACCAUGUGACCUACCCAUGAG (((....((((((((((((((..((((..(((.....)))..))))...((....))((((((.....)))).))...))))))))))))))..)))....((((.(......))))).. ( -38.60) >sp_pn.0 1692470 120 + 2038615/2200-2320 CGGCGAGUUACGUUAUGAUGCGAGGUUAAGUUGAAGAGACGGAGCCGUAGGGAAACCGAGUCUGAAUAGGGCACCUUAGUAUCAUGACGUAGACCCGAAACCAUGUGACCUACCCAUGAG (((....(((((((((((((((((((............(((....))).((....))..((((.....))))))))).))))))))))))))..)))....((((.(......))))).. ( -39.80) >sp_mu.0 1873490 120 + 2030921/2200-2320 CGGCGAGUUACGUUUACGUGCGAGGUUAAGUUGAAGAGACGGAGCCGUAGGGAAACCGAGUCUGAAAAGGGCGGUUAAGUACGUAGAUGUAGACCCGAAACCAAGUGACCUACCCAUGAG (((....((((((((((((((..((((..(((.....)))..)))).......(((((..(((....))).)))))..))))))))))))))..)))....................... ( -37.50) >consensus CGGCGAGUUACGAUUUGAUGCAAGGUUAAGCAGAAAAGACGGAGCCGUAGCGAAACCGAGUCUGAAUAGGGCG_UUUAGUAUUUGGUCGUAGACCCGAAACCAGGUGACCUACCCAUGAG (((....((((((((.(((((..((((..(((.....)))..))))...((....))..((((.....))))......))))).))))))))..)))....................... (-36.04 = -32.30 + -3.74) # Strand winner: reverse (0.99)
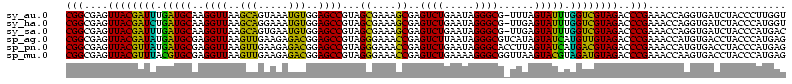
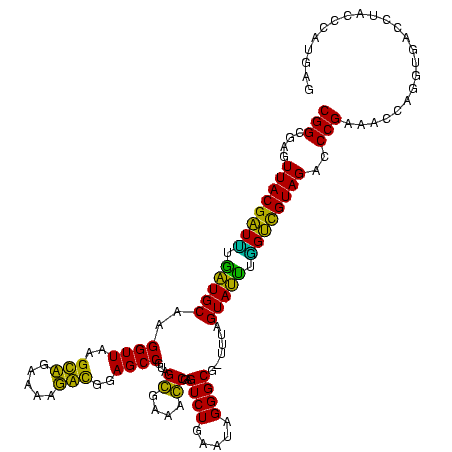
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.56 |
| Mean single sequence MFE | -38.69 |
| Consensus MFE | -37.35 |
| Energy contribution | -36.63 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.97 |
| SVM decision value | 4.42 |
| SVM RNA-class probability | 0.999894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/2320-2440 GUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAUAGAACCUGAAACCGUGUGCUUACAAGUAGUCAGAGCCCGUUAAUGGGUGAUGGCGUGCCUUUUGUAGAAUGAAC ...((....))(((((((...(((((((((....)))).((........))...........))))).......((((..(((((....)))))..))))..)))))))........... ( -39.50) >sy_ha.0 1576954 120 - 2685015/2320-2440 GUACCGUGAGGGAAAGGUGAAAAGUACCCCGGAAGGGGAGUGAAAGAGAACUUGAAACCGUGUGCUUACAAGUAGUCAGAGCCCGUUAAUAGGUGAUGGCGUGCCUUUUGUAGAAUGAAC ...((....))(((((((...(((((((((....)))((((........))))........)))))).......((((..(((........)))..))))..)))))))........... ( -36.00) >sy_sa.0 1551999 120 - 2516575/2320-2440 GUACCGUGAGGGAAAGGUGAAAAGUACCCCGGAAGGGGAGUGAAAUAGAACUUGAAACCGUGUGCUUACAAGUAGUCAGAGCCCGUUAAUGGGUGAUGGCGUGCCUUUUGUAGAAUGAAC ...((....))(((((((...(((((((((....)))((((........))))........)))))).......((((..(((((....)))))..))))..)))))))........... ( -40.00) >sp_ag.0 2133292 120 - 2160267/2320-2440 GUACCGUGAGGGAAAGGUGAAAAGCACCCCGGGAGGGGAGUGAAAUAGAACCUGAAACCGUGUGCCUACAACAAGUUCGAGCCCGUUAAUGGGUGAGAGCGUGCCUUUUGUAGAAUGAAC (((((....)((..((((.....((.((((....)))).))........))))....))..))))((((((((.((((..(((((....)))))..)))).))....))))))....... ( -38.12) >sp_pn.0 1692470 120 + 2038615/2320-2440 GUACCGUGAGGGAAAGGUGAAAAGCACCCCGGGAGGGGAGUGAAAUAGAACCUGAAACCGUGUGCCUACAACAAGUUCGAGCCCGUUAAUGGGUGAGAGCGUGCCUUUUGUAGAAUGAAC (((((....)((..((((.....((.((((....)))).))........))))....))..))))((((((((.((((..(((((....)))))..)))).))....))))))....... ( -38.12) >sp_mu.0 1873490 120 + 2030921/2320-2440 GUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAACCUGAAACCGUGUGCUUACAAGAAGUUCGAGCCCGUUAAUGGGUGAGAGCGUGCCUUUUGUAGAAUGAAC ...((....))(((((((...(((((((((....)))).((...((.....))...))....))))).......((((..(((((....)))))..))))..)))))))........... ( -40.40) >consensus GUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAUAGAACCUGAAACCGUGUGCUUACAAGAAGUCAGAGCCCGUUAAUGGGUGAGAGCGUGCCUUUUGUAGAAUGAAC ...((....))(((((((...(((((((((....)))).((........))...........))))).......((((..(((((....)))))..))))..)))))))........... (-37.35 = -36.63 + -0.72) # Strand winner: reverse (1.00)
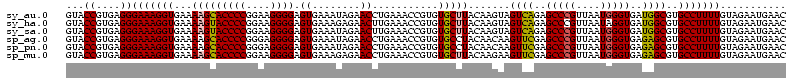
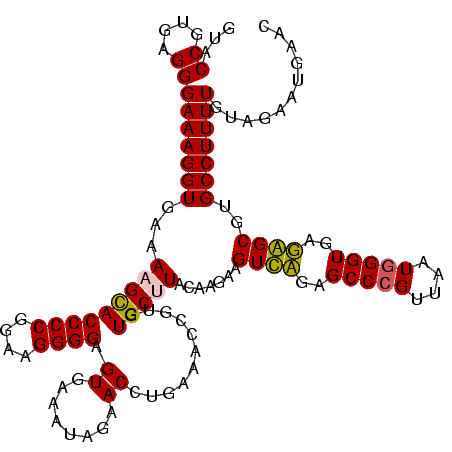
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.22 |
| Mean single sequence MFE | -38.42 |
| Consensus MFE | -38.47 |
| Energy contribution | -37.33 |
| Covariance contribution | -1.14 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.19 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/2400-2520 CGGAACACGUGAAAUUCCGUCGGAAUCUGGGAGGACCAUCUCCUAAGGCUAAAUACUCUCUAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAG .((((.........))))(((((....((((((......))))))..((((.........))))..))))).((..(((....((....))....))).....)).((((....)))).. ( -36.80) >sy_ha.0 1576954 120 - 2685015/2400-2520 CGGAGCACGUGAAAUUCCGUCGGAAUCUGGGAGGACCAUCUCCUAAGGCUAAAUACUCUCUAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGUACCCCGGAAGGGGAG .((((.........))))(((((....((((((......))))))..((((.........))))..))))).....(((....((....))....)))........((((....)))).. ( -35.50) >sy_sa.0 1551999 120 - 2516575/2400-2520 CGGAACACGAGAAAUUCCGUCGGAAUCUGGGAGGACCAUCUCCCAAGGCUAAAUACUCUCUAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGUACCCCGGAAGGGGAG (((..(((((((.(((((...))))).((((((......))))))...........))))..))).))).......(((....((....))....)))........((((....)))).. ( -38.70) >sp_ag.0 2133292 120 - 2160267/2400-2520 CGAGACACGCGAAAUCUCGUCGGAAUCUGGGAGGACCAUCUCCCAACCCUAAAUACUCUCUAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGGAGGGGAG .((((.........))))(((((....((((((......))))))........((((....)))).))))).((..(((....((....))....))).....)).((((....)))).. ( -37.60) >sp_pn.0 1692470 120 + 2038615/2400-2520 CGGGACACGUGAAAUCCCGUCGGAAUCUGGGAGGACCAUCUCCCAACCCUAAAUACUCCCUAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGGAGGGGAG .((((.........))))(((((....((((((......))))))........((((....)))).))))).((..(((....((....))....))).....)).((((....)))).. ( -39.70) >sp_mu.0 1873490 120 + 2030921/2400-2520 CGGGACACGAGGAAUCCCGUCGGAAUCUGGGAGGCCCAUCUCCCAACCCUAAAUACUCCCUAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAG (((..(((.(((..(((....)))...(((((((....))))))).............))).))).)))...((..(((....((....))....))).....)).((((....)))).. ( -42.20) >consensus CGGAACACGUGAAAUCCCGUCGGAAUCUGGGAGGACCAUCUCCCAACCCUAAAUACUCUCUAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAG .((((.........))))(((((....((((((......))))))........((((....)))).))))).....(((....((....))....)))........((((....)))).. (-38.47 = -37.33 + -1.14) # Strand winner: reverse (0.99)

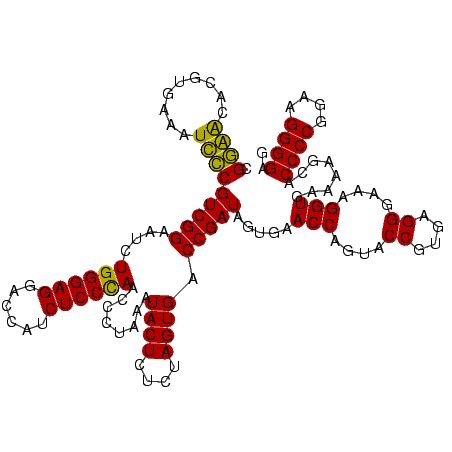
| Location | 1,977,118 – 1,977,237 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.35 |
| Mean single sequence MFE | -32.68 |
| Consensus MFE | -28.17 |
| Energy contribution | -24.62 |
| Covariance contribution | -3.55 |
| Combinations/Pair | 1.54 |
| Mean z-score | -0.77 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.569120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 119 + 2809422/2480-2600 CAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCGAAAAUGUUGUCUCUCUUGAGUGG-AUCCUGAGUACGACGGAACACGUGAAAUUCCGUCGGAAUCUGGGAGGACCAUC (((((....)))))..(((((((.(..((((.((........))...))))..)))).))))((((-.((((((...((((((((.........))))))))...)).))))...)))). ( -32.50) >sy_ha.0 1576954 119 - 2685015/2480-2600 CAUCUGGAAAGUUGAAUCAAAGAAGGUAAUAAUCCUGUAGUUGAAAAUAUAUUCUCUCUUGAGUGG-AUCCUGAGUACGACGGAGCACGUGAAAUUCCGUCGGAAUCUGGGAGGACCAUC ((.((....)).))..((((...(((.......)))....)))).........(((....)))(((-.((((((...((((((((.........))))))))...)).))))...))).. ( -28.40) >sy_sa.0 1551999 119 - 2516575/2480-2600 CAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCGAAAGUUUGUUCACUCUUGAGUGG-AUCCUGAGUACGACGGAACACGAGAAAUUCCGUCGGAAUCUGGGAGGACCAUC (((((....)))))..........(((.....(((((((.(((...(((..((((....))))..)-))..))).)))(((((((.........))))))).......))))..)))... ( -34.10) >sp_ag.0 2133292 120 - 2160267/2480-2600 UACCUGGGAAGGUAAGCCAAAGAGAGUAACAGCCUCGUAUUUAAAAUGAUAGAUUAACCUUAGCAGUAUCCUGAGUACGGCGAGACACGCGAAAUCUCGUCGGAAUCUGGGAGGACCAUC ...(((..(((((........(((.((....))))).((((......)))).....)))))..)))..((((((...((((((((.........))))))))...)).))))........ ( -29.00) >sp_pn.0 1692470 119 + 2038615/2480-2600 GAUUUGGGAAGAUCAGCCAAAGAGAGUAAUAGCCUCGUAUUUAAAAUAGU-CUUUGUACUUAGCAGUAUCCUGAGUACGGCGGGACACGUGAAAUCCCGUCGGAAUCUGGGAGGACCAUC (((((....))))).......(((.((....)))))............((-((((((((((((.......))))))))(((((((.........))))))).........)))))).... ( -31.60) >sp_mu.0 1873490 119 + 2030921/2480-2600 ACUCUGGGAAGAGUCGCCAGAGAGAGUAAGAGCCUCGUAUUUGAAAU-UCACUUGAUGCCAAGCAGGAUCCUGAGUACGGCGGGACACGAGGAAUCCCGUCGGAAUCUGGGAGGCCCAUC .((((((((....)).)))))).......(.(((((.........((-((.((((....))))..))))((.((...((((((((.........))))))))...)).))))))).)... ( -40.50) >consensus CAUCUGGAAAGAUGAACCAAAGAAAGUAAUAACCCCGUAGUUGAAAUUAUACUCUCUCCUGAGCAG_AUCCUGAGUACGACGGAACACGUGAAAUCCCGUCGGAAUCUGGGAGGACCAUC (((((....)))))..((.....(((.......)))................................((((((...((((((((.........))))))))...)).))))))...... (-28.17 = -24.62 + -3.55) # Strand winner: reverse (1.00)

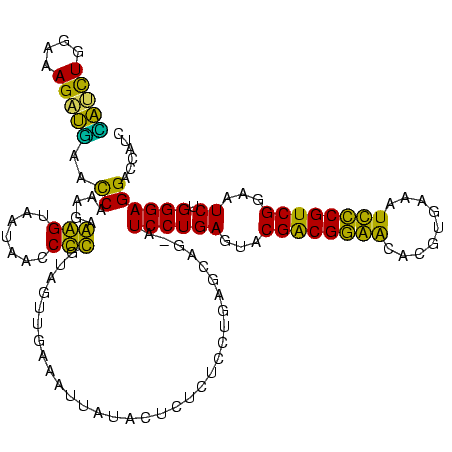

| Location | 1,977,118 – 1,977,237 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.06 |
| Mean single sequence MFE | -29.24 |
| Consensus MFE | -12.26 |
| Energy contribution | -9.46 |
| Covariance contribution | -2.80 |
| Combinations/Pair | 1.70 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.42 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 119 + 2809422/2520-2640 ACACUCUAUACGGAGUUACAAAGGACGACAUUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCGAAAAUGUUGUCUCUCUUGAGUGG-AUCCUGAGUACGA .(((((.....((((.......((((((((((.(((((.((((((....)))))).)).....(((.......)))...)))...)))))))))))))).))))).-............. ( -35.01) >sy_ha.0 1576954 119 - 2685015/2520-2640 ACACUCUAUACGGAGUUACAAAGGAAUAUGUUAGACGAAUCAUCUGGAAAGUUGAAUCAAAGAAGGUAAUAAUCCUGUAGUUGAAAAUAUAUUCUCUCUUGAGUGG-AUCCUGAGUACGA .(((((.....((((.......((((((((((.(((((.(((.((....)).))).)).....(((.......)))...)))...)))))))))))))).))))).-............. ( -25.21) >sy_sa.0 1551999 119 - 2516575/2520-2640 ACACUCUAUACGGAGUUACAAAAGAACAGACUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAGUCGAAAGUUUGUUCACUCUUGAGUGG-AUCCUGAGUACGA .(((((.....(((((.......(((((((((.(((((.((((((....)))))).)).....(((.......)))...)))...)))))))))))))).))))).-............. ( -35.31) >sp_ag.0 2133292 113 - 2160267/2520-2640 AC-UGCGAUAUAGGAUUAAUCAUUA------UAGAAGAAUUACCUGGGAAGGUAAGCCAAAGAGAGUAACAGCCUCGUAUUUAAAAUGAUAGAUUAACCUUAGCAGUAUCCUGAGUACGG ((-(((.....(((.((((((((((------(....(..((((((....))))))..)...(((.((....))))).........))))).)))))))))..)))))............. ( -26.70) >sp_pn.0 1692470 112 + 2038615/2520-2640 AC-UGCAAUGUGGACUCAAAGAUUA------UAGAAGAAUGAUUUGGGAAGAUCAGCCAAAGAGAGUAAUAGCCUCGUAUUUAAAAUAGU-CUUUGUACUUAGCAGUAUCCUGAGUACGG ((-(((...(((....(((((((((------(....(..((((((....))))))..)...(((.((....))))).........)))))-))))))))...)))))............. ( -24.40) >sp_mu.0 1873490 112 + 2030921/2520-2640 AC-UGCGAUAAAGCAGCCAAGGGAA------UAGAAGAAGACUCUGGGAAGAGUCGCCAGAGAGAGUAAGAGCCUCGUAUUUGAAAU-UCACUUGAUGCCAAGCAGGAUCCUGAGUACGG .(-(((......(((..((((.(((------(.......((((((....))))))..((((..(((.......)))...))))..))-)).)))).)))...)))).............. ( -28.80) >consensus AC_CGCUAUACGGAGUUAAAAAGGA______UAGAAGAAUCAUCUGGAAAGAUGAACCAAAGAAAGUAAUAACCCCGUAGUUGAAAUUAUACUCUCUCCUGAGCAG_AUCCUGAGUACGA ...(((......)))........................((((((....)))))).((.....(((.......)))(((.(((........(((......)))........))).))))) (-12.26 = -9.46 + -2.80) # Strand winner: reverse (0.99)
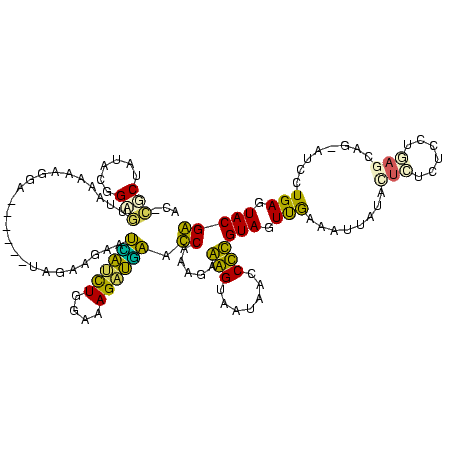
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.77 |
| Mean single sequence MFE | -27.38 |
| Consensus MFE | -23.01 |
| Energy contribution | -18.72 |
| Covariance contribution | -4.30 |
| Combinations/Pair | 1.68 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/2560-2680 CUACAGGAUUAUUACCUUCUUUGAUUCAUCUUUCCAGAUGAUUCGUCUAAUGUCGUCCUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAACAAGCAAGCUUGUUGGUUUGGGCUCUU .((((((((.((.........(((.((((((....)))))).)))......)).))))..))))((((......))))((((.......((((((((....))))))))...)))).... ( -30.16) >sy_ha.0 1576954 120 - 2685015/2560-2680 CUACAGGAUUAUUACCUUCUUUGAUUCAACUUUCCAGAUGAUUCGUCUAACAUAUUCCUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAAUAAGCAAGCUUAUUGGUUUGGGCUCUU .(((((((.(((.........(((.(((.((....)).))).)))......))).)))..))))((((......))))((((.......((((((((....))))))))...)))).... ( -23.06) >sy_sa.0 1551999 120 - 2516575/2560-2680 CUACAGGAUUAUUACCUUCUUUGAUUCAUCUUUCCAGAUGAUUCGUCUAGUCUGUUCUUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAACAAGCAAGCUCGUUGGUUUGGGCUCUU .(((((((..((...((....(((.((((((....)))))).)))...))...))..)))))))((((......))))((((.......(((((.((....)).)))))...)))).... ( -26.60) >sp_ag.0 2133292 113 - 2160267/2560-2680 AUACGAGGCUGUUACUCUCUUUGGCUUACCUUCCCAGGUAAUUCUUCUA------UAAUGAUUAAUCCUAUAUCGCA-GUCCUACAACCCCGAAGUGUAAACACUUCGGUUUGCCCUCCU ....((((((((..........(..((((((....))))))..).....------....(((.........))))))-)..........((((((((....)))))))).....)))).. ( -25.40) >sp_pn.0 1692470 113 + 2038615/2560-2680 AUACGAGGCUAUUACUCUCUUUGGCUGAUCUUCCCAAAUCAUUCUUCUA------UAAUCUUUGAGUCCACAUUGCA-GUCCUACAACCCCGAAGAGUAAACUCUUCGGUUUGCCCUCCU ....(((((.............(((((...........(((........------.......)))..........))-)))........((((((((....))))))))...).)))).. ( -23.26) >sp_mu.0 1873490 113 + 2030921/2560-2680 AUACGAGGCUCUUACUCUCUCUGGCGACUCUUCCCAGAGUCUUCUUCUA------UUCCCUUGGCUGCUUUAUCGCA-GUCCUACAACCCCAGAGUGUAAACACUCUGGUUUGCCCUCCU ....(((.......))).....(((((((((....))))))........------.......((((((......)))-)))........((((((((....))))))))...)))..... ( -35.80) >consensus AUACAAGACUAUUACCCUCUUUGACUCAUCUUCCCAGAUGAUUCGUCUA______UCCUUUGUAACUCCGUAUAGAA_GUCCUACAACCCCAAAAAGCAAACUCUUUGGUUUGCCCUCCU ....(((.......))).....((.((((((....)))))).))........................................(((..((((((((....)))))))).)))....... (-23.01 = -18.72 + -4.30)
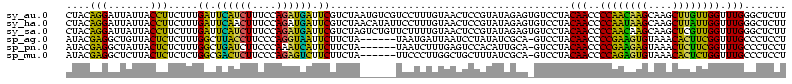
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.77 |
| Mean single sequence MFE | -33.07 |
| Consensus MFE | -27.89 |
| Energy contribution | -23.03 |
| Covariance contribution | -4.85 |
| Combinations/Pair | 1.84 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.84 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/2560-2680 AAGAGCCCAAACCAACAAGCUUGCUUGUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGACGACAUUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAG .(((((((((.((((((((....))))))))..)))..))...)))).(((((((((((...(.....).......((.((((((....)))))).)).......)))))..))).))). ( -32.90) >sy_ha.0 1576954 120 - 2685015/2560-2680 AAGAGCCCAAACCAAUAAGCUUGCUUAUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGAAUAUGUUAGACGAAUCAUCUGGAAAGUUGAAUCAAAGAAGGUAAUAAUCCUGUAG ...(((((....(((((((....)))))))))))).(((((.(((((....)))))..........(((.((....((.(((.((....)).))).))....)).)))....)))))... ( -26.40) >sy_sa.0 1551999 120 - 2516575/2560-2680 AAGAGCCCAAACCAACGAGCUUGCUUGUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAAGAACAGACUAGACGAAUCAUCUGGAAAGAUGAAUCAAAGAAGGUAAUAAUCCUGUAG .(((((((((.((((((((....))))))))..)))..))...)))).(((((((((((...((......))....((.((((((....)))))).)).......)))))..))).))). ( -32.20) >sp_ag.0 2133292 113 - 2160267/2560-2680 AGGAGGGCAAACCGAAGUGUUUACACUUCGGGGUUGUAGGAC-UGCGAUAUAGGAUUAAUCAUUA------UAGAAGAAUUACCUGGGAAGGUAAGCCAAAGAGAGUAACAGCCUCGUAU ..((((((((.((((((((....))))))))..)))).((.(-(....(((((((....)).)))------))..))..((((((....)))))).))..............)))).... ( -32.30) >sp_pn.0 1692470 113 + 2038615/2560-2680 AGGAGGGCAAACCGAAGAGUUUACUCUUCGGGGUUGUAGGAC-UGCAAUGUGGACUCAAAGAUUA------UAGAAGAAUGAUUUGGGAAGAUCAGCCAAAGAGAGUAAUAGCCUCGUAU ..((((((...((((((((....)))))))).((((((....-))))))))..((((..((((((------(......))))))).((........)).....)))).....)))).... ( -31.70) >sp_mu.0 1873490 113 + 2030921/2560-2680 AGGAGGGCAAACCAGAGUGUUUACACUCUGGGGUUGUAGGAC-UGCGAUAAAGCAGCCAAGGGAA------UAGAAGAAGACUCUGGGAAGAGUCGCCAGAGAGAGUAAGAGCCUCGUAU ..((((((((.((((((((....))))))))..)))).((.(-(((......)))))).......------..........((((((((....)).))))))..........)))).... ( -42.90) >consensus AAGAGCCCAAACCAAAGAGCUUACUCUUUGGGGUUGUAGGAC_CGCUAUACGGAGUUAAAAAGGA______UAGAAGAAUCAUCUGGAAAGAUGAACCAAAGAAAGUAAUAACCCCGUAG .((........((((((((....))))))))((((((......(((......)))........................((((((....)))))).............)))))))).... (-27.89 = -23.03 + -4.85) # Strand winner: reverse (0.99)

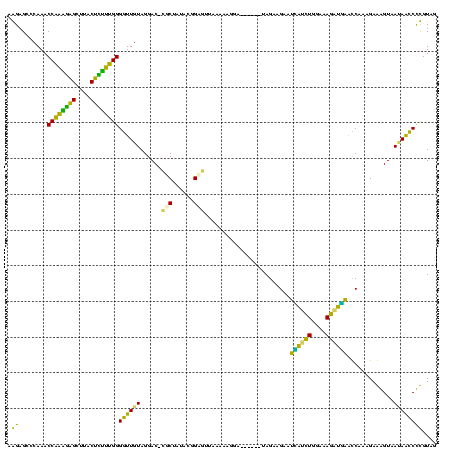
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.11 |
| Mean single sequence MFE | -28.55 |
| Consensus MFE | -22.37 |
| Energy contribution | -19.18 |
| Covariance contribution | -3.19 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.981068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/2600-2720 AUUCGUCUAAUGUCGUCCUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAACAAGCAAGCUUGUUGGUUUGGGCUCUUCCCGUUUCGCUCGCCGCUACUAAGGGAAUCGAAUUUUCUU (((((.........((((.(((((((((......))))....)))))..((((((((....))))))))...))))..(((((.....((.....))......))))).)))))...... ( -30.90) >sy_ha.0 1576954 120 - 2685015/2600-2720 AUUCGUCUAACAUAUUCCUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAAUAAGCAAGCUUAUUGGUUUGGGCUCUUCCCGUUUCGCUCGCCGCUACUCAGGGAAUCGAUUUUUCUU ..((((((((.........(((((((((......))))....)))))..((((((((....)))))))).))))))..(((((.....((.....))......)))))..))........ ( -26.30) >sy_sa.0 1551999 120 - 2516575/2600-2720 AUUCGUCUAGUCUGUUCUUUUGUAACUCCGUAUAGAGUGUCCUACAACCCCAACAAGCAAGCUCGUUGGUUUGGGCUCUUCCCGUUUCGCUCGCCGCUACUCAGGGAAUCGAUUUUUCUU ....(...((((.(((((((.(((....((....(((((....((....(((((.((....)).)))))...(((.....)))))..)))))..)).)))..))))))).))))...).. ( -25.00) >sp_ag.0 2133292 113 - 2160267/2600-2720 AUUCUUCUA------UAAUGAUUAAUCCUAUAUCGCA-GUCCUACAACCCCGAAGUGUAAACACUUCGGUUUGCCCUCCUGCCGUUUCGCUCGCCGCUACUAAGGCAAUCGCUUUUGCUU .........------....((((...........(((-(.....(((..((((((((....)))))))).))).....))))..........(((........)))))))((....)).. ( -26.50) >sp_pn.0 1692470 113 + 2038615/2600-2720 AUUCUUCUA------UAAUCUUUGAGUCCACAUUGCA-GUCCUACAACCCCGAAGAGUAAACUCUUCGGUUUGCCCUCCUGCCGUUUCGCUCGCCGCUACUAAGGCAAUCGCUUUUGCUU .........------........((((....((.(((-(.....(((..((((((((....)))))))).))).....)))).))...))))(((........)))....((....)).. ( -28.50) >sp_mu.0 1873490 113 + 2030921/2600-2720 CUUCUUCUA------UUCCCUUGGCUGCUUUAUCGCA-GUCCUACAACCCCAGAGUGUAAACACUCUGGUUUGCCCUCCUGCCUCUUCGCUCGCCGCUACUGAGGCAAUCGCUCUUGCUU .........------.......((((((......)))-)))........((((((((....))))))))..........((((((...((.....))....))))))...((....)).. ( -34.10) >consensus AUUCGUCUA______UCCUUUGUAACUCCGUAUAGAA_GUCCUACAACCCCAAAAAGCAAACUCUUUGGUUUGCCCUCCUCCCGUUUCGCUCGCCGCUACUAAGGCAAUCGAUUUUGCUU ..................................(((.......(((..((((((((....)))))))).)))......((((.....((.....))......))))........))).. (-22.37 = -19.18 + -3.19)
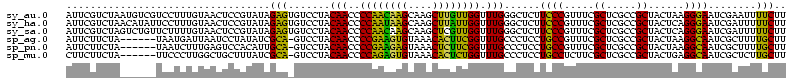
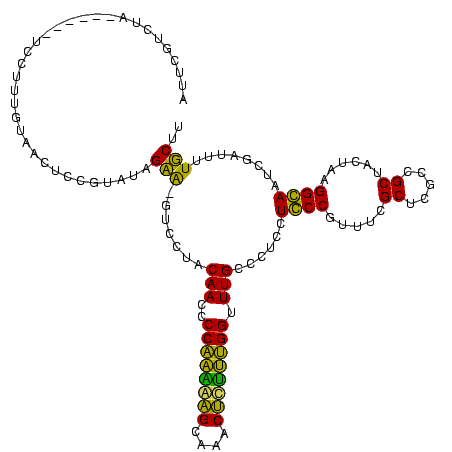
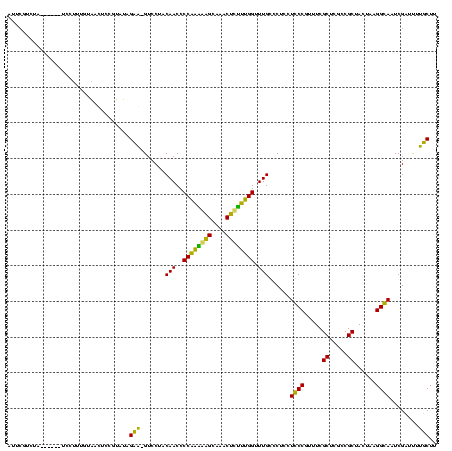
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.11 |
| Mean single sequence MFE | -33.45 |
| Consensus MFE | -26.86 |
| Energy contribution | -23.37 |
| Covariance contribution | -3.49 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/2600-2720 AAGAAAAUUCGAUUCCCUUAGUAGCGGCGAGCGAAACGGGAAGAGCCCAAACCAACAAGCUUGCUUGUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGACGACAUUAGACGAAU ......(((((....((((.(((((..(((((.....(((.....)))...((((((((....)))))))).)))(((((....))))).))..))))).)))).(.......).))))) ( -33.00) >sy_ha.0 1576954 120 - 2685015/2600-2720 AAGAAAAAUCGAUUCCCUGAGUAGCGGCGAGCGAAACGGGAAGAGCCCAAACCAAUAAGCUUGCUUAUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGAAUAUGUUAGACGAAU ........(((.(((((......((.....)).....)))))....((...((((((((....))))))))..(((((....(((((....)))))))))).))...........))).. ( -29.10) >sy_sa.0 1551999 120 - 2516575/2600-2720 AAGAAAAAUCGAUUCCCUGAGUAGCGGCGAGCGAAACGGGAAGAGCCCAAACCAACGAGCUUGCUUGUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAAGAACAGACUAGACGAAU ..........((((((..((((..(.((((.(.....(((.....)))...((((((((....))))))))).))))..)..)))).....))))))....................... ( -30.40) >sp_ag.0 2133292 113 - 2160267/2600-2720 AAGCAAAAGCGAUUGCCUUAGUAGCGGCGAGCGAAACGGCAGGAGGGCAAACCGAAGUGUUUACACUUCGGGGUUGUAGGAC-UGCGAUAUAGGAUUAAUCAUUA------UAGAAGAAU .........((.(((((........))))).)).....((((....((((.((((((((....))))))))..))))....)-)))..(((((((....)).)))------))....... ( -32.60) >sp_pn.0 1692470 113 + 2038615/2600-2720 AAGCAAAAGCGAUUGCCUUAGUAGCGGCGAGCGAAACGGCAGGAGGGCAAACCGAAGAGUUUACUCUUCGGGGUUGUAGGAC-UGCAAUGUGGACUCAAAGAUUA------UAGAAGAAU ..((....))(((((((........)))((((.(....((((....((((.((((((((....))))))))..))))....)-)))....).).)))...)))).------......... ( -34.80) >sp_mu.0 1873490 113 + 2030921/2600-2720 AAGCAAGAGCGAUUGCCUCAGUAGCGGCGAGCGAAGAGGCAGGAGGGCAAACCAGAGUGUUUACACUCUGGGGUUGUAGGAC-UGCGAUAAAGCAGCCAAGGGAA------UAGAAGAAG ..((....))..(((((((....((.....))...)))))))....((((.((((((((....))))))))..)))).((.(-(((......)))))).......------......... ( -40.80) >consensus AAGAAAAAGCGAUUCCCUUAGUAGCGGCGAGCGAAACGGCAAGAGCCCAAACCAAAGAGCUUACUCUUUGGGGUUGUAGGAC_CGCUAUACGGAGUUAAAAAGGA______UAGAAGAAU ..((.....(..(((((......((.....)).....)))))..)..(((.((((((((....))))))))..)))........)).................................. (-26.86 = -23.37 + -3.49) # Strand winner: reverse (0.99)
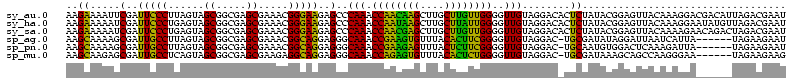
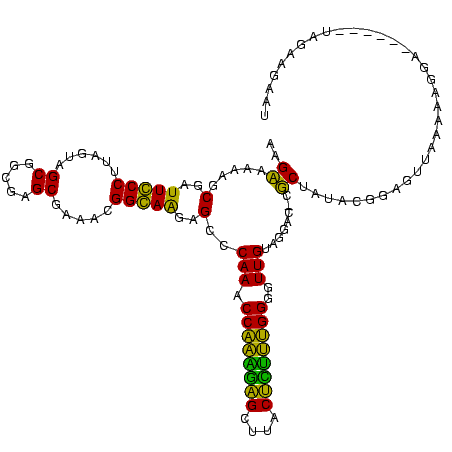

| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.50 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -28.78 |
| Energy contribution | -24.90 |
| Covariance contribution | -3.88 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/2640-2760 CCUACAACCCCAACAAGCAAGCUUGUUGGUUUGGGCUCUUCCCGUUUCGCUCGCCGCUACUAAGGGAAUCGAAUUUUCUUUUUCUUCCUCCGGGUACUAAGAUGUUUCAGUUCUCCGGGU ((((.....((((((((....))))))))..)))).....((((....((.....))......((((((.(((...((((......((....))....))))...))).)))))))))). ( -31.10) >sy_ha.0 1576954 120 - 2685015/2640-2760 CCUACAACCCCAAUAAGCAAGCUUAUUGGUUUGGGCUCUUCCCGUUUCGCUCGCCGCUACUCAGGGAAUCGAUUUUUCUUUCUCUUCCUCCGGGUACUAAGAUGUUUCAGUUCUCCGGGU ((((.....((((((((....))))))))..)))).....((((....((.....))......((((((.((..(.((((......((....))....)))).)..)).)))))))))). ( -28.00) >sy_sa.0 1551999 120 - 2516575/2640-2760 CCUACAACCCCAACAAGCAAGCUCGUUGGUUUGGGCUCUUCCCGUUUCGCUCGCCGCUACUCAGGGAAUCGAUUUUUCUUUCUCUUCCUGCGGGUACUAAGAUGUUUCAGUUCUCCGCGU .........(((((.((....)).)))))...(((.....)))....(((...((((.....((((((..((....)).))))))....)))).......((....))........))). ( -25.70) >sp_ag.0 2133292 120 - 2160267/2640-2760 CCUACAACCCCGAAGUGUAAACACUUCGGUUUGCCCUCCUGCCGUUUCGCUCGCCGCUACUAAGGCAAUCGCUUUUGCUUUCUCUUCCUGCAGCUACUUAGAUGUUUCAGUUCACUGCGU .........((((((((....)))))))).(((((.....((.((.......)).))......)))))..((....))...........((((..(((..((....)))))...)))).. ( -27.10) >sp_pn.0 1692470 120 + 2038615/2640-2760 CCUACAACCCCGAAGAGUAAACUCUUCGGUUUGCCCUCCUGCCGUUUCGCUCGCCGCUACUAAGGCAAUCGCUUUUGCUUUCUCUUCCUGCAGCUACUUAGAUGUUUCAGUUCACUGCGU .........((((((((....)))))))).(((((.....((.((.......)).))......)))))..((....))...........((((..(((..((....)))))...)))).. ( -27.30) >sp_mu.0 1873490 120 + 2030921/2640-2760 CCUACAACCCCAGAGUGUAAACACUCUGGUUUGCCCUCCUGCCUCUUCGCUCGCCGCUACUGAGGCAAUCGCUCUUGCUUUCUCUUCCUGCAGCUACUGAGAUGUUUCAGUUCACUGCGU .........((((((((....))))))))..........((((((...((.....))....))))))...((....))...........((((..((((((....))))))...)))).. ( -36.80) >consensus CCUACAACCCCAAAAAGCAAACUCUUUGGUUUGCCCUCCUCCCGUUUCGCUCGCCGCUACUAAGGCAAUCGAUUUUGCUUUCUCUUCCUGCAGCUACUAAGAUGUUUCAGUUCACCGCGU ....(((..((((((((....)))))))).)))......((((.....((.....))......))))...((....))..........(((((..(((..((....)))))...))))). (-28.78 = -24.90 + -3.88)
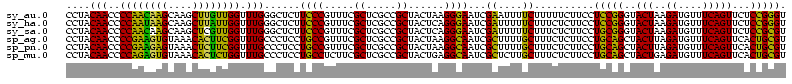
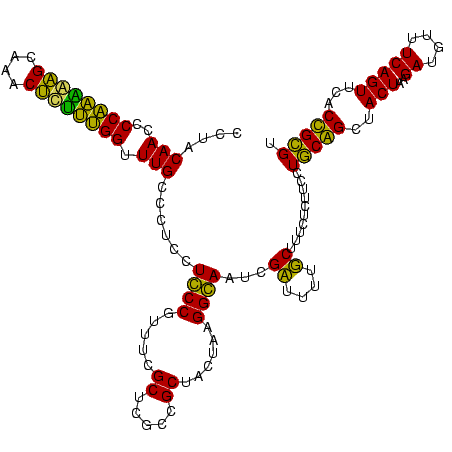
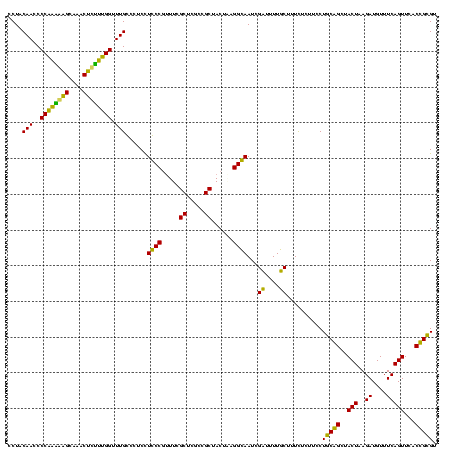
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.50 |
| Mean single sequence MFE | -37.38 |
| Consensus MFE | -34.47 |
| Energy contribution | -30.28 |
| Covariance contribution | -4.19 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/2640-2760 ACCCGGAGAACUGAAACAUCUUAGUACCCGGAGGAAGAAAAAGAAAAUUCGAUUCCCUUAGUAGCGGCGAGCGAAACGGGAAGAGCCCAAACCAACAAGCUUGCUUGUUGGGGUUGUAGG ..((((.(.(((((......))))).))))).(((.(((........)))...))).......(((((.........(((.....)))...((((((((....)))))))).)))))... ( -33.90) >sy_ha.0 1576954 120 - 2685015/2640-2760 ACCCGGAGAACUGAAACAUCUUAGUACCCGGAGGAAGAGAAAGAAAAAUCGAUUCCCUGAGUAGCGGCGAGCGAAACGGGAAGAGCCCAAACCAAUAAGCUUGCUUAUUGGGGUUGUAGG ..((((.(.(((((......))))).))))).((((..((........))..)))).......(((((.........(((.....)))...((((((((....)))))))).)))))... ( -34.50) >sy_sa.0 1551999 120 - 2516575/2640-2760 ACGCGGAGAACUGAAACAUCUUAGUACCCGCAGGAAGAGAAAGAAAAAUCGAUUCCCUGAGUAGCGGCGAGCGAAACGGGAAGAGCCCAAACCAACGAGCUUGCUUGUUGGGGUUGUAGG ..((((.(.(((((......))))).))))).((((..((........))..)))).......(((((.........(((.....)))...((((((((....)))))))).)))))... ( -36.80) >sp_ag.0 2133292 120 - 2160267/2640-2760 ACGCAGUGAACUGAAACAUCUAAGUAGCUGCAGGAAGAGAAAGCAAAAGCGAUUGCCUUAGUAGCGGCGAGCGAAACGGCAGGAGGGCAAACCGAAGUGUUUACACUUCGGGGUUGUAGG ..(((((..(((..........))).)))))...........((.....((.(((((........))))).)).....))......((((.((((((((....))))))))..))))... ( -36.90) >sp_pn.0 1692470 120 + 2038615/2640-2760 ACGCAGUGAACUGAAACAUCUAAGUAGCUGCAGGAAGAGAAAGCAAAAGCGAUUGCCUUAGUAGCGGCGAGCGAAACGGCAGGAGGGCAAACCGAAGAGUUUACUCUUCGGGGUUGUAGG ..(((((..(((..........))).)))))...........((.....((.(((((........))))).)).....))......((((.((((((((....))))))))..))))... ( -37.10) >sp_mu.0 1873490 120 + 2030921/2640-2760 ACGCAGUGAACUGAAACAUCUCAGUAGCUGCAGGAAGAGAAAGCAAGAGCGAUUGCCUCAGUAGCGGCGAGCGAAGAGGCAGGAGGGCAAACCAGAGUGUUUACACUCUGGGGUUGUAGG ..(((((..(((((......))))).)))))...........((....))..(((((((....((.....))...)))))))....((((.((((((((....))))))))..))))... ( -45.10) >consensus ACGCAGAGAACUGAAACAUCUUAGUACCCGCAGGAAGAGAAAGAAAAAGCGAUUCCCUUAGUAGCGGCGAGCGAAACGGCAAGAGCCCAAACCAAAGAGCUUACUCUUUGGGGUUGUAGG ..((((...(((((......)))))..))))...........((....))..(((((......((.....)).....))))).....(((.((((((((....))))))))..))).... (-34.47 = -30.28 + -4.19) # Strand winner: reverse (1.00)
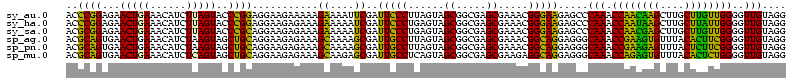
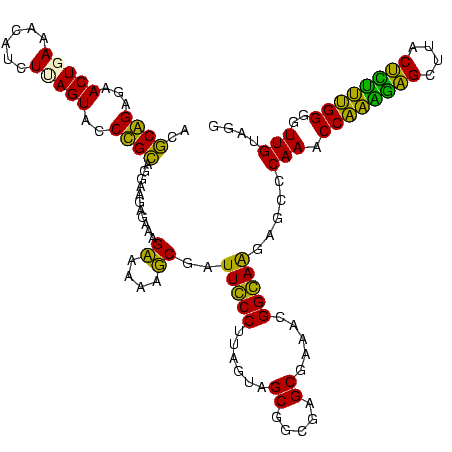
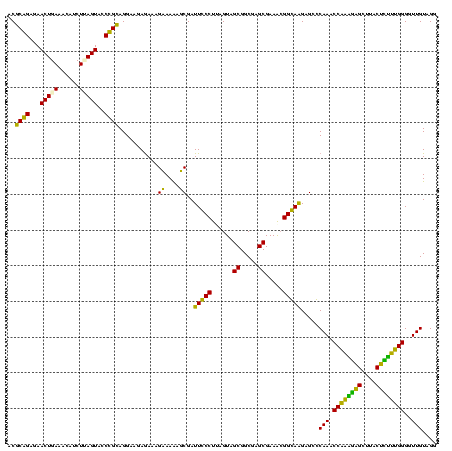
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.44 |
| Mean single sequence MFE | -26.98 |
| Consensus MFE | -22.38 |
| Energy contribution | -19.30 |
| Covariance contribution | -3.08 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/2720-2840 UUUCUUCCUCCGGGUACUAAGAUGUUUCAGUUCUCCGGGUGUGCCUUCUGAUAUGCUAUGUAUUCACAUAUCGAUAACAUGACAUAACUCAUGCUGGGUUUCCCCAUUCGGAAAUCUCUG ........(((((((.....((....))........(((...((((..(((((((...........)))))))....(((((......)))))..))))...))))))))))........ ( -25.00) >sy_ha.0 1576954 120 - 2685015/2720-2840 UCUCUUCCUCCGGGUACUAAGAUGUUUCAGUUCUCCGGGUCUGCCUUCUGACAAGCUAUGAAUUCACUUGUCGAUAACACGACAUAACUCGUGCUGGGUUCCCCCAUUCGGAAAUCUCUG ........(((((((.....((....))........(((...((((..(((((((...((....)))))))))....(((((......)))))..))))...))))))))))........ ( -28.50) >sy_sa.0 1551999 120 - 2516575/2720-2840 UCUCUUCCUGCGGGUACUAAGAUGUUUCAGUUCUCCGCGUCUGCCUUCAGAUAUGCUAUGUAUUCACAUAUCGAUAACACGACAUAACUCGUGCUGGGUUUCCCCAUUCGGAAAUCUCUG .........(((((.(((..((....))))).).))))(((((....))))).((.(((((....))))).))....(((((......)))))..((((((((......))))))))... ( -26.40) >sp_ag.0 2133292 113 - 2160267/2720-2840 UCUCUUCCUGCAGCUACUUAGAUGUUUCAGUUCACUGCGUCUUCCUUCUCAUAUCCUUAA----CAGAUAUGGAUACUA-GUCAUUAAC--UAGUGGGUUCCCCCAUUCGGACAUCUCUG .........((((..(((..((....)))))...))))((((....((.((((((.....----..)))))))).((((-((.....))--))))(((....)))....))))....... ( -23.40) >sp_pn.0 1692470 113 + 2038615/2720-2840 UCUCUUCCUGCAGCUACUUAGAUGUUUCAGUUCACUGCGUCUUCCUCCUCACAUCCUUAA----CAGAUGUGGGUAACA-GGUAUUACC--UGUUGGGUUCCCCCAUUCGGAAAUCCCUG .........((((..(((..((....)))))...))))...((((..((((((((.....----..)))))))).((((-((.....))--))))(((....)))....))))....... ( -29.90) >sp_mu.0 1873490 113 + 2030921/2720-2840 UCUCUUCCUGCAGCUACUGAGAUGUUUCAGUUCACUGCGUCUUCCUUCUCAUAACCUUAA----CAGUUAUGGAUAACA-GGCAUUACC--UGUUGGGUUCCCCCAUUCGGAUAUCUCUG .........((((..((((((....))))))...))))....(((.((.((((((.....----..)))))))).((((-((.....))--))))(((....)))....)))........ ( -28.70) >consensus UCUCUUCCUGCAGCUACUAAGAUGUUUCAGUUCACCGCGUCUGCCUUCUCAUAUCCUAAA____CACAUAUCGAUAACA_GACAUAACC__UGCUGGGUUCCCCCAUUCGGAAAUCUCUG .....(((.((((..(((..((....)))))...))))(((.....((.((((((...........))))))))......)))...........((((....))))...)))........ (-22.38 = -19.30 + -3.08)
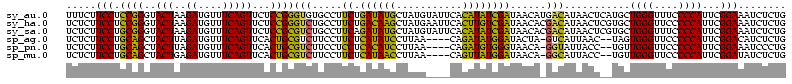
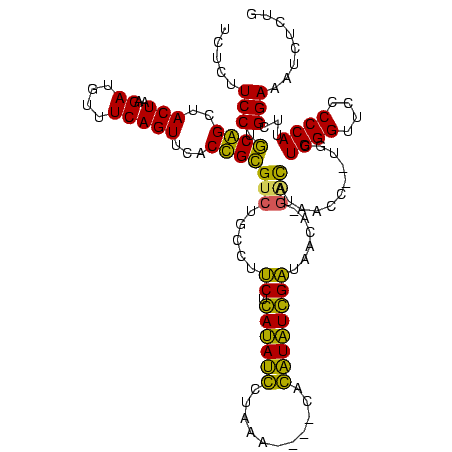
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.44 |
| Mean single sequence MFE | -37.23 |
| Consensus MFE | -31.19 |
| Energy contribution | -27.97 |
| Covariance contribution | -3.23 |
| Combinations/Pair | 1.39 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969212 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/2720-2840 CAGAGAUUUCCGAAUGGGGAAACCCAGCAUGAGUUAUGUCAUGUUAUCGAUAUGUGAAUACAUAGCAUAUCAGAAGGCACACCCGGAGAACUGAAACAUCUUAGUACCCGGAGGAAGAAA ......(((((((..(((....)))(((((((......))))))).))(((((((.........)))))))...........((((.(.(((((......))))).))))).)))))... ( -36.70) >sy_ha.0 1576954 120 - 2685015/2720-2840 CAGAGAUUUCCGAAUGGGGGAACCCAGCACGAGUUAUGUCGUGUUAUCGACAAGUGAAUUCAUAGCUUGUCAGAAGGCAGACCCGGAGAACUGAAACAUCUUAGUACCCGGAGGAAGAGA ......(((((((..(((....)))(((((((......))))))).))(((((((.........)))))))...........((((.(.(((((......))))).))))).)))))... ( -38.30) >sy_sa.0 1551999 120 - 2516575/2720-2840 CAGAGAUUUCCGAAUGGGGAAACCCAGCACGAGUUAUGUCGUGUUAUCGAUAUGUGAAUACAUAGCAUAUCUGAAGGCAGACGCGGAGAACUGAAACAUCUUAGUACCCGCAGGAAGAGA ......(((((...((((....))))...........(((.(((..(((((((((.........))))))).))..))))))((((.(.(((((......))))).))))).)))))... ( -39.10) >sp_ag.0 2133292 113 - 2160267/2720-2840 CAGAGAUGUCCGAAUGGGGGAACCCACUA--GUUAAUGAC-UAGUAUCCAUAUCUG----UUAAGGAUAUGAGAAGGAAGACGCAGUGAACUGAAACAUCUAAGUAGCUGCAGGAAGAGA ........(((....(((....)))((((--(((...)))-)))).(((((((((.----....))))))).)).)))....(((((..(((..........))).)))))......... ( -31.80) >sp_pn.0 1692470 113 + 2038615/2720-2840 CAGGGAUUUCCGAAUGGGGGAACCCAACA--GGUAAUACC-UGUUACCCACAUCUG----UUAAGGAUGUGAGGAGGAAGACGCAGUGAACUGAAACAUCUAAGUAGCUGCAGGAAGAGA ......(((((....(((....)))((((--((.....))-)))).(((((((((.----....))))))).)).)))))..(((((..(((..........))).)))))......... ( -39.10) >sp_mu.0 1873490 113 + 2030921/2720-2840 CAGAGAUAUCCGAAUGGGGGAACCCAACA--GGUAAUGCC-UGUUAUCCAUAACUG----UUAAGGUUAUGAGAAGGAAGACGCAGUGAACUGAAACAUCUCAGUAGCUGCAGGAAGAGA ........(((....(((....)))((((--((.....))-)))).(((((((((.----....))))))).)).)))....(((((..(((((......))))).)))))......... ( -38.40) >consensus CAGAGAUUUCCGAAUGGGGGAACCCAGCA__AGUAAUGUC_UGUUAUCCAUAUCUG____CAAAGCAUAUCAGAAGGAAGACGCAGAGAACUGAAACAUCUUAGUACCCGCAGGAAGAGA ......(((((....(((....)))(((((..........))))).(((((((((.........))))))).))........((((...(((((......)))))..)))).)))))... (-31.19 = -27.97 + -3.23) # Strand winner: reverse (1.00)
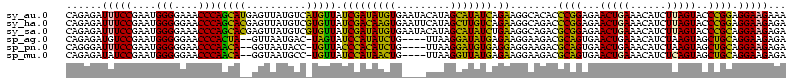
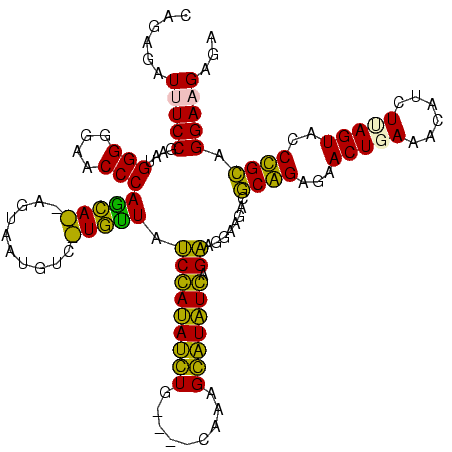

| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.90 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -18.89 |
| Energy contribution | -16.45 |
| Covariance contribution | -2.44 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/2760-2880 GUGCCUUCUGAUAUGCUAUGUAUUCACAUAUCGAUAACAUGACAUAACUCAUGCUGGGUUUCCCCAUUCGGAAAUCUCUGGAUCAAAGCUUACUUACAGCUCCCCAAAGCAUAUCGUCGU .........((((((((.((.(((((...........(((((......)))))..((((((((......)))))))).))))))).((((.......))))......))))))))..... ( -26.80) >sy_ha.0 1576954 120 - 2685015/2760-2880 CUGCCUUCUGACAAGCUAUGAAUUCACUUGUCGAUAACACGACAUAACUCGUGCUGGGUUCCCCCAUUCGGAAAUCUCUGGAUCAACGCUUACUUACAGCUACCCAAAGCAUAUCGUCGU .........((((((...((....))))))))((((.(((((......))))).(((((......(((((((....)))))))....(((.......))).))))).....))))..... ( -26.20) >sy_sa.0 1551999 120 - 2516575/2760-2880 CUGCCUUCAGAUAUGCUAUGUAUUCACAUAUCGAUAACACGACAUAACUCGUGCUGGGUUUCCCCAUUCGGAAAUCUCUGGAUCAAAGCUUACUUACAGCUCCCCAAAGCAUAUCGUCGU .........((((((((.((.(((((...........(((((......)))))..((((((((......)))))))).))))))).((((.......))))......))))))))..... ( -28.40) >sp_ag.0 2133292 113 - 2160267/2760-2880 CUUCCUUCUCAUAUCCUUAA----CAGAUAUGGAUACUA-GUCAUUAAC--UAGUGGGUUCCCCCAUUCGGACAUCUCUGGAUCAGCGCUUACUUACAGCUCCCCAAAGCAUUUCGUCGU ......((.((((((.....----..)))))))).((((-((.....))--))))(((....)))...((((...((.(((...(((...........)))..))).))...)))).... ( -21.00) >sp_pn.0 1692470 113 + 2038615/2760-2880 CUUCCUCCUCACAUCCUUAA----CAGAUGUGGGUAACA-GGUAUUACC--UGUUGGGUUCCCCCAUUCGGAAAUCCCUGGAUCAUCGCUUACUUACAGCUACCCAAGGCAUAUCGUCGU .......((((((((.....----..)))))))).((((-((.....))--))))((((......((((((......))))))....(((.......))).))))..(((.....))).. ( -28.90) >sp_mu.0 1873490 113 + 2030921/2760-2880 CUUCCUUCUCAUAACCUUAA----CAGUUAUGGAUAACA-GGCAUUACC--UGUUGGGUUCCCCCAUUCGGAUAUCUCUGGAUCAAGGCUUACUUACAGCUCCCCAAAGCAUGUCGUCGU .........((((((.....----..))))))(((((((-((.....))--))))(((....))).....(((((((.(((.....((((.......))))..))).)).)))))))).. ( -24.70) >consensus CUGCCUUCUCAUAUCCUAAA____CACAUAUCGAUAACA_GACAUAACC__UGCUGGGUUCCCCCAUUCGGAAAUCUCUGGAUCAACGCUUACUUACAGCUCCCCAAAGCAUAUCGUCGU ......((.((((((...........))))))))......(((...........((((....))))(((((......)))))................(((......))).....))).. (-18.89 = -16.45 + -2.44)
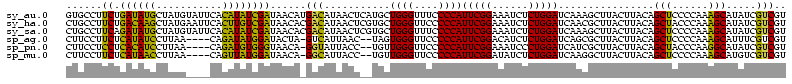
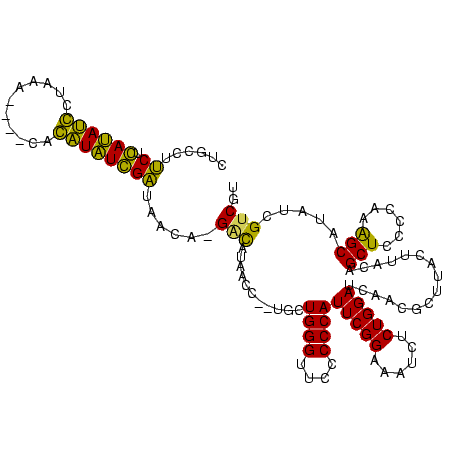
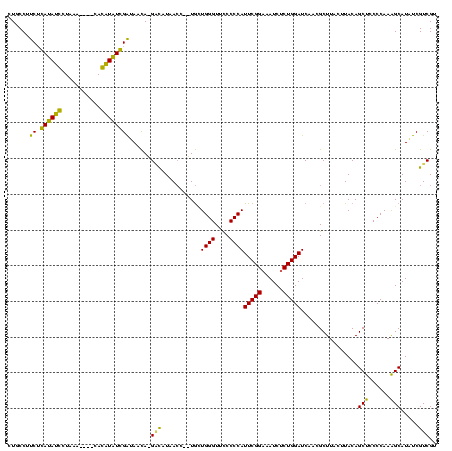
| Location | 1,977,118 – 1,977,238 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.90 |
| Mean single sequence MFE | -38.52 |
| Consensus MFE | -30.61 |
| Energy contribution | -27.47 |
| Covariance contribution | -3.14 |
| Combinations/Pair | 1.34 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 120 + 2809422/2760-2880 ACGACGAUAUGCUUUGGGGAGCUGUAAGUAAGCUUUGAUCCAGAGAUUUCCGAAUGGGGAAACCCAGCAUGAGUUAUGUCAUGUUAUCGAUAUGUGAAUACAUAGCAUAUCAGAAGGCAC .....((((((((((((((((((.......)))))...))))))......(((..(((....)))(((((((......))))))).))).(((((....))))))))))))......... ( -37.50) >sy_ha.0 1576954 120 - 2685015/2760-2880 ACGACGAUAUGCUUUGGGUAGCUGUAAGUAAGCGUUGAUCCAGAGAUUUCCGAAUGGGGGAACCCAGCACGAGUUAUGUCGUGUUAUCGACAAGUGAAUUCAUAGCUUGUCAGAAGGCAG ...((((((((((((((((.(((.......)))....)))))))).....((..((((....))))...))...))))))))((..(((((((((.........))))))).))..)).. ( -40.20) >sy_sa.0 1551999 120 - 2516575/2760-2880 ACGACGAUAUGCUUUGGGGAGCUGUAAGUAAGCUUUGAUCCAGAGAUUUCCGAAUGGGGAAACCCAGCACGAGUUAUGUCGUGUUAUCGAUAUGUGAAUACAUAGCAUAUCUGAAGGCAG ...((((((((((((((((((((.......)))))...))))))).....((..((((....))))...))...))))))))((..(((((((((.........))))))).))..)).. ( -39.60) >sp_ag.0 2133292 113 - 2160267/2760-2880 ACGACGAAAUGCUUUGGGGAGCUGUAAGUAAGCGCUGAUCCAGAGAUGUCCGAAUGGGGGAACCCACUA--GUUAAUGAC-UAGUAUCCAUAUCUG----UUAAGGAUAUGAGAAGGAAG ...........(((((((.(((.((......)))))..)))))))...(((....(((....)))((((--(((...)))-)))).(((((((((.----....))))))).)).))).. ( -34.60) >sp_pn.0 1692470 113 + 2038615/2760-2880 ACGACGAUAUGCCUUGGGUAGCUGUAAGUAAGCGAUGAUCCAGGGAUUUCCGAAUGGGGGAACCCAACA--GGUAAUACC-UGUUACCCACAUCUG----UUAAGGAUGUGAGGAGGAAG ...........((((((((.(((.......)))....))))))))..((((....(((....)))((((--((.....))-)))).(((((((((.----....))))))).)).)))). ( -44.40) >sp_mu.0 1873490 113 + 2030921/2760-2880 ACGACGACAUGCUUUGGGGAGCUGUAAGUAAGCCUUGAUCCAGAGAUAUCCGAAUGGGGGAACCCAACA--GGUAAUGCC-UGUUAUCCAUAACUG----UUAAGGUUAUGAGAAGGAAG ...........(((((((..(((.......))).....)))))))...(((....(((....)))((((--((.....))-)))).(((((((((.----....))))))).)).))).. ( -34.80) >consensus ACGACGAUAUGCUUUGGGGAGCUGUAAGUAAGCGUUGAUCCAGAGAUUUCCGAAUGGGGGAACCCAGCA__AGUAAUGUC_UGUUAUCCAUAUCUG____CAAAGCAUAUCAGAAGGAAG ....((.....(((((((..(((.......))).....))))))).....))...(((....)))(((((..........))))).(((((((((.........))))))).))...... (-30.61 = -27.47 + -3.14) # Strand winner: reverse (1.00)

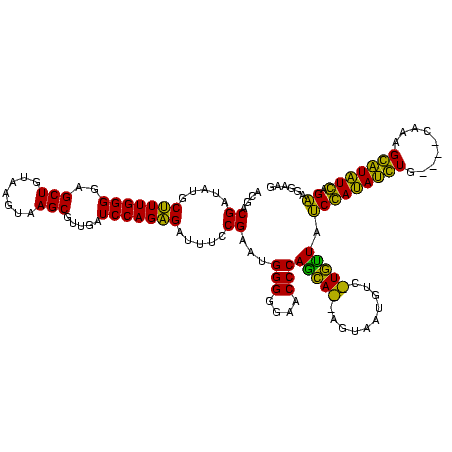
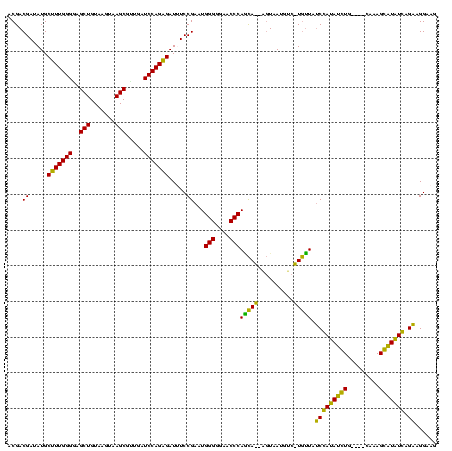
| Location | 1,977,118 – 1,977,208 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.71 |
| Mean single sequence MFE | -32.63 |
| Consensus MFE | -27.70 |
| Energy contribution | -27.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 90 + 2809422/3156-3276 C----------------AAUGUACA---------AA-UAAU----GGUGGAGACUAGCGGGAUCGAACCGCUGACCUCCUGCGUGCAAAGCAGGCGCUCUCCCAGCUGAGCUAAGCCCCC .----------------........---------..-....----(((((((..((((((.......))))))..)))).((.(((...))).))((((........))))...)))... ( -29.80) >sy_ha.0 1576954 94 - 2685015/3156-3276 C----------------AAUGUACA---------AA-UAAUUAAUGGUGGAGACUAGCGGGAUCGAACCGCUGACCUCCUGCGUGCAAAGCAGGCGCUCUCCCAGCUGAGCUAAGCCCCC .----------------........---------..-........(((((((..((((((.......))))))..)))).((.(((...))).))((((........))))...)))... ( -29.80) >sy_sa.0 1551999 95 - 2516575/3156-3276 C----------------AAUGUACA---------AAAUUUUUCAUGGUGGAGACUAGCGGGAUCGAACCGCUGACCUCCUGCGUGCAAAGCAGGCGCUCUCCCAUCUGAGCUAAGCCCCC .----------------........---------...........(((((((..((((((.......))))))..)))).((.(((...))).))((((........))))...)))... ( -29.80) >sp_ag.0 2133292 102 - 2160267/3156-3276 C----------------AAUGAACA--AUUUGAACCUUUCGAUUCAAUGGAGCCUAGCGGGAUCGAACCGCUGACCUCCUGCGUGCAAAGCAGGCGCUCUCCCAGCUGAGCUAAGGCCCC .----------------........--..(((((.(....).))))).((.(((((((((.......))))).....(((((.......))))).((((........))))..)))))). ( -30.80) >sp_pn.0 1692470 104 + 2038615/3156-3276 C----------------AAUGUGCAUUACUUGGUGAUCUCUCACCAAUGGAGCCUAGCGGGAUCGAACCGCUGACCUCCUGCGUGCAAAGCAGGCGCUCUCCCAGCUGAGCUAAGGCCCC .----------------............(((((((....))))))).((.(((((((((.......))))).....(((((.......))))).((((........))))..)))))). ( -37.40) >sp_mu.0 1873490 120 + 2030921/3156-3276 CUACAGAAGUAUUCGCAAGCGAACCGUCUGUUAGUAUCCUGUUUUAAUGGAGCCUAGCGGGAUCGAACCGCUGACCUCCUGCGUGCAAAGCAGGCGCUCUCCCAGCUGAGCUAAGGCCCC (((((((....((((....))))...)))).)))......((((((..((((..((((((.......))))))..)))).((.(((...))).))((((........))))))))))... ( -38.20) >consensus C________________AAUGUACA_________AAUUUAUUAAUAAUGGAGACUAGCGGGAUCGAACCGCUGACCUCCUGCGUGCAAAGCAGGCGCUCUCCCAGCUGAGCUAAGCCCCC ................................................((((..((((((.......))))))..)))).((.(((...))).))((((........))))......... (-27.70 = -27.70 + -0.00)

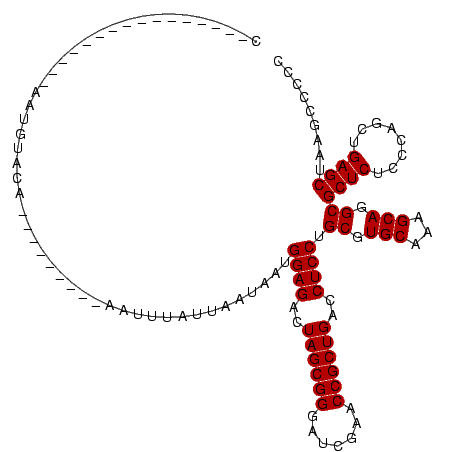
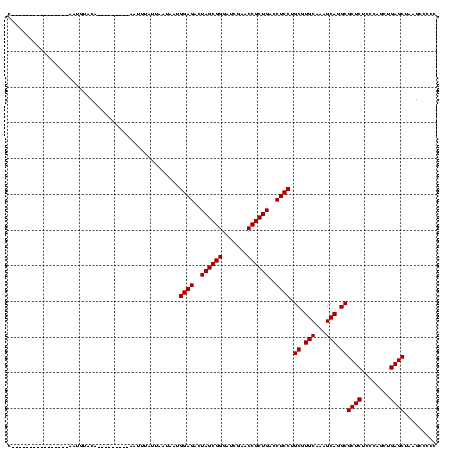
| Location | 1,977,118 – 1,977,208 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.71 |
| Mean single sequence MFE | -35.97 |
| Consensus MFE | -31.55 |
| Energy contribution | -31.05 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.05 |
| Mean z-score | -0.85 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1977118 90 + 2809422/3156-3276 GGGGGCUUAGCUCAGCUGGGAGAGCGCCUGCUUUGCACGCAGGAGGUCAGCGGUUCGAUCCCGCUAGUCUCCACC----AUUA-UU---------UGUACAUU----------------G ((((((...)))).((..(.(((((....))))).)..)).(((((..(((((.......)))))..))))).))----....-..---------........----------------. ( -29.90) >sy_ha.0 1576954 94 - 2685015/3156-3276 GGGGGCUUAGCUCAGCUGGGAGAGCGCCUGCUUUGCACGCAGGAGGUCAGCGGUUCGAUCCCGCUAGUCUCCACCAUUAAUUA-UU---------UGUACAUU----------------G ((((((...)))).((..(.(((((....))))).)..)).(((((..(((((.......)))))..))))).))........-..---------........----------------. ( -29.90) >sy_sa.0 1551999 95 - 2516575/3156-3276 GGGGGCUUAGCUCAGAUGGGAGAGCGCCUGCUUUGCACGCAGGAGGUCAGCGGUUCGAUCCCGCUAGUCUCCACCAUGAAAAAUUU---------UGUACAUU----------------G (((((((.(((...(((.(((..((.(((((.......)))))......))..))).)))..))))))))))..............---------........----------------. ( -29.70) >sp_ag.0 2133292 102 - 2160267/3156-3276 GGGGCCUUAGCUCAGCUGGGAGAGCGCCUGCUUUGCACGCAGGAGGUCAGCGGUUCGAUCCCGCUAGGCUCCAUUGAAUCGAAAGGUUCAAAU--UGUUCAUU----------------G (((((((..((((........)))).(((((.......))))).....(((((.......)))))))))))).(((((((....)))))))..--........----------------. ( -41.80) >sp_pn.0 1692470 104 + 2038615/3156-3276 GGGGCCUUAGCUCAGCUGGGAGAGCGCCUGCUUUGCACGCAGGAGGUCAGCGGUUCGAUCCCGCUAGGCUCCAUUGGUGAGAGAUCACCAAGUAAUGCACAUU----------------G (((((((..((((........)))).(((((.......))))).....(((((.......)))))))))))).(((((((....)))))))............----------------. ( -41.50) >sp_mu.0 1873490 120 + 2030921/3156-3276 GGGGCCUUAGCUCAGCUGGGAGAGCGCCUGCUUUGCACGCAGGAGGUCAGCGGUUCGAUCCCGCUAGGCUCCAUUAAAACAGGAUACUAACAGACGGUUCGCUUGCGAAUACUUCUGUAG (((((((..((((........)))).(((((.......))))).....(((((.......)))))))))))).................(((((..(((((....)))))...))))).. ( -43.00) >consensus GGGGCCUUAGCUCAGCUGGGAGAGCGCCUGCUUUGCACGCAGGAGGUCAGCGGUUCGAUCCCGCUAGGCUCCACCAAAAAAAAAUU_________UGUACAUU________________G (((((((..((((........)))).(((((.......))))).....(((((.......))))))))))))................................................ (-31.55 = -31.05 + -0.50) # Strand winner: reverse (0.98)

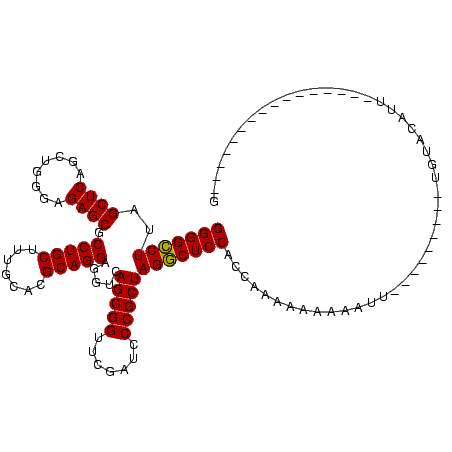
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:55:31 2006