| Sequence ID | sy_sa.0 |
|---|---|
| Location | 1,825,917 – 1,826,026 |
| Length | 109 |
| Max. P | 0.978679 |

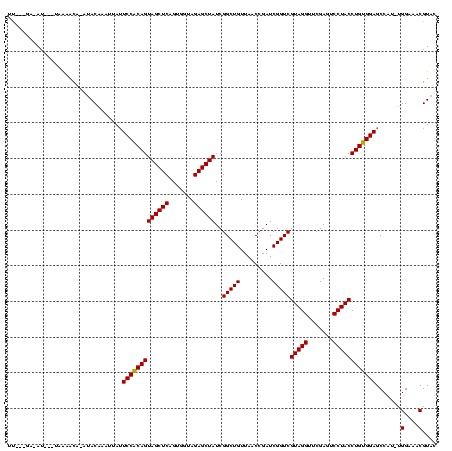
| Location | 1,825,917 – 1,826,025 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.27 |
| Mean single sequence MFE | -35.87 |
| Consensus MFE | -32.08 |
| Energy contribution | -32.30 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 1825917 108 + 2516575/80-200 GUACGUUUCCA-AUGGCUCCACAGGUAGGACUCGAACCUACGACCGAUCGGUUAACAGCCGAUAGCUCUACCACUGAGCUACUGUGGAAUAAUGGGU---UGCUCUA---AU--C---AA ....((.((((-.((..((((((((((((.......))))).....((((((.....))))))(((((.......))))).))))))).)).)))).---.))....---..--.---.. ( -36.00) >sy_au.0 1042231 113 + 2809422/80-200 GUACGUUUCCA-AUGGCUCCACAGGUAGGACUCGAACCUACGACCGAUCGGUUAACAGCCGAUAGCUCUACCACUGAGCUACUGUGGAUUAAUUUGUAUGUGUUUGA---AUAUC---AA .(((((...((-(..(.((((((((((((.......))))).....((((((.....))))))(((((.......))))).))))))).)...))).))))).....---.....---.. ( -33.60) >sy_ha.0 710687 120 - 2685015/80-200 GUACGUUUCCAUAUGGCUCCGCAGGUAGGAUUCGAACCUACGACCGAUCGGUUAACAGCCGAUAGCUCUACCACUGAGCUACUGCGGAAUAAUAUGUAUUUAUAUAAUGUAUCUCUAUGA (((((((..(((((...((((((((((((.......))))).....((((((.....))))))(((((.......))))).)))))))...))))).........)))))))........ ( -38.00) >consensus GUACGUUUCCA_AUGGCUCCACAGGUAGGACUCGAACCUACGACCGAUCGGUUAACAGCCGAUAGCUCUACCACUGAGCUACUGUGGAAUAAUAUGUAU_UGUUUAA___AU_UC___AA ((((....((....)).((((((((((((.......))))).....((((((.....))))))(((((.......))))).))))))).......))))..................... (-32.08 = -32.30 + 0.22)

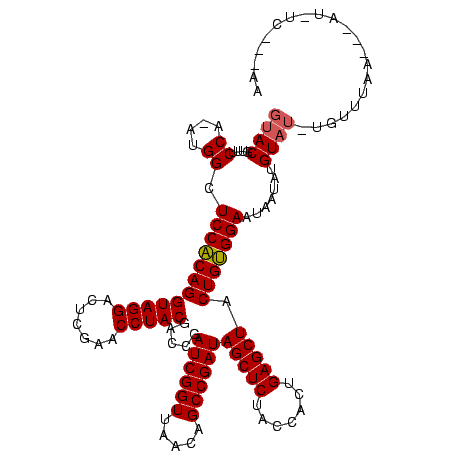
| Location | 1,825,917 – 1,826,025 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.27 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -30.48 |
| Energy contribution | -30.03 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832700 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 1825917 108 + 2516575/80-200 UU---G--AU---UAGAGCA---ACCCAUUAUUCCACAGUAGCUCAGUGGUAGAGCUAUCGGCUGUUAACCGAUCGGUCGUAGGUUCGAGUCCUACCUGUGGAGCCAU-UGGAAACGUAC ..---.--..---.......---..(((...((((((((((((((.......))))))..(((((.........)))))(((((.......)))))))))))))....-)))........ ( -31.50) >sy_au.0 1042231 113 + 2809422/80-200 UU---GAUAU---UCAAACACAUACAAAUUAAUCCACAGUAGCUCAGUGGUAGAGCUAUCGGCUGUUAACCGAUCGGUCGUAGGUUCGAGUCCUACCUGUGGAGCCAU-UGGAAACGUAC ..---.....---.........(((.......(((((((((((((.......))))))..(((((.........)))))(((((.......)))))))))))).....-.(....)))). ( -30.80) >sy_ha.0 710687 120 - 2685015/80-200 UCAUAGAGAUACAUUAUAUAAAUACAUAUUAUUCCGCAGUAGCUCAGUGGUAGAGCUAUCGGCUGUUAACCGAUCGGUCGUAGGUUCGAAUCCUACCUGCGGAGCCAUAUGGAAACGUAC ........................(((((..((((((((((((((.......))))))..(((((.........)))))(((((.......)))))))))))))..)))))......... ( -34.50) >consensus UU___GA_AU___UAAAACA_AUACAAAUUAUUCCACAGUAGCUCAGUGGUAGAGCUAUCGGCUGUUAACCGAUCGGUCGUAGGUUCGAGUCCUACCUGUGGAGCCAU_UGGAAACGUAC ................................(((((((((((((.......))))))..(((((.........)))))(((((.......)))))))))))).......(....).... (-30.48 = -30.03 + -0.44) # Strand winner: forward (0.83)

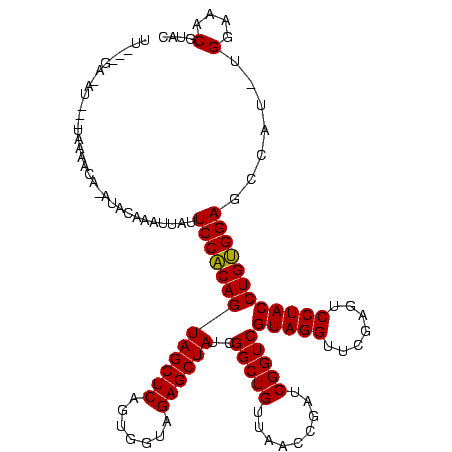
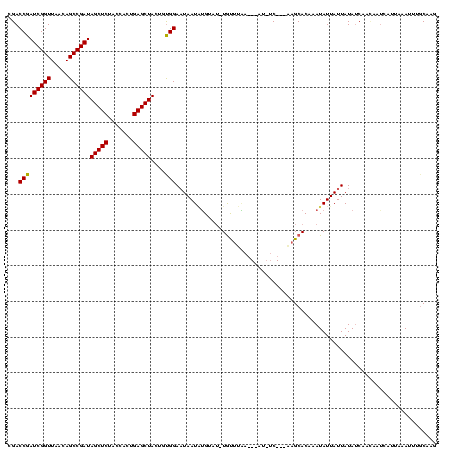
| Location | 1,825,917 – 1,826,026 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.38 |
| Mean single sequence MFE | -26.37 |
| Consensus MFE | -16.82 |
| Energy contribution | -16.60 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
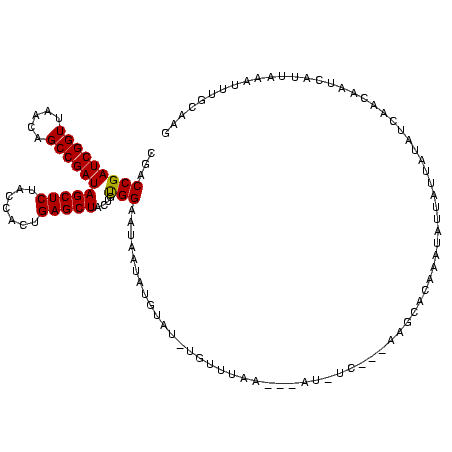
>sy_sa.0 1825917 109 + 2516575/120-240 CGACCGAUCGGUUAACAGCCGAUAGCUCUACCACUGAGCUACUGUGGAAUAAUGGGU---UGCUCUA---AU--C---AAGCACAAAUAUUAUUAUAUCAACUCUCGUUAAAUUUUCAAG ((((((((((((.....))))))(((((.......)))))....)))(((((((.((---.(((...---..--.---.)))))...)))))))..........)))............. ( -22.90) >sy_au.0 1042231 113 + 2809422/120-240 CGACCGAUCGGUUAACAGCCGAUAGCUCUACCACUGAGCUACUGUGGAUUAAUUUGUAUGUGUUUGA---AUAUC---AAGCACAAAUAUUAUUAUAACAAGAUUAAGUUAAUUUGCAA- ......((((((.....))))))(((((.......)))))..((..((((((((((..((((((((.---....)---))))))).(((....)))........))))))))))..)).- ( -33.30) >sy_ha.0 710687 120 - 2685015/120-240 CGACCGAUCGGUUAACAGCCGAUAGCUCUACCACUGAGCUACUGCGGAAUAAUAUGUAUUUAUAUAAUGUAUCUCUAUGAGCACAAAUAUUAUUAUAUCAAUAAACUUAAAAUUUGCAAG .(((((((((((.....))))))(((((.......)))))....)))((((((((((.((((((...........)))))).)))..)))))))...))..................... ( -22.90) >consensus CGACCGAUCGGUUAACAGCCGAUAGCUCUACCACUGAGCUACUGUGGAAUAAUAUGUAU_UGUUUAA___AU_UC___AAGCACAAAUAUUAUUAUAUCAACAAUCAUUAAAUUUGCAAG ...(((((((((.....))))))(((((.......)))))....)))......................................................................... (-16.82 = -16.60 + -0.22)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:54:49 2006