
| Sequence ID | sy_au.0 |
|---|---|
| Location | 1,976,242 – 1,976,362 |
| Length | 120 |
| Max. P | 0.998549 |

| Location | 1,976,242 – 1,976,356 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.13 |
| Mean single sequence MFE | -30.98 |
| Consensus MFE | -27.18 |
| Energy contribution | -27.50 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1976242 114 + 2809422/0-120 UCGGGAAGACAGGAUUCGAACCUGCGACCCCUUGGUCCCAAACCAAGUGCUCUACCAAGCUGAGCUACUUCCCGUAU------AAUUAACGCGCCCGAUAGGAGUCGAACCCAUAACCUC .(((((((.((((.......))))......((((((.....)))))).((((.........))))..)))))))...------......((.((((....)).))))............. ( -29.70) >sy_ha.0 1576076 113 - 2685015/0-120 UCGGGAAGACAGGAUUCGAACCUGCGACCCCUUGGUCCCAAACCAAGUGCUCUACCAAACUGAGCUACUUCCCGUAA------GAG-AACGCGCCCGAUAGGAGUCGAACCCAUAACCUC .(((((((.((((.......))))......((((((.....)))))).((((.........))))..)))))))...------...-..((.((((....)).))))............. ( -29.70) >sy_sa.0 1551116 113 - 2516575/0-120 UCGGGAAGACAGGAUUCGAACCUGCGACCCCUUGGUCCCAAACCAAGUGCUCUACCAAGCUGAGCUACUUCCCGUA-------AAUUAGUGCGCCCAAUAGGGGUCGAACCUAUAACCUC ..(((....((((.......))))(((((((((((.........((((((((.........)))).))))..((((-------......)))).)))..))))))))..)))........ ( -33.10) >sp_ag.0 2132345 112 - 2160267/0-120 UCGGGAAGACAGGAUUCGAACCUGCGACACCUUGGUCCCAAACCAAGUACUCUACCAAGCUGAGCUACUUCCCGAA--------AAAUAUGCACCCUAGAGGAGUCGAACCUCUAACCGC ((((((((.((((.......))))......((((((.....))))))..(((.........)))...)))))))).--------............((((((.......))))))..... ( -31.20) >sp_pn.0 1691504 120 + 2038615/0-120 UCGGGAAGACAGGAUUCGAACCUGCGACACCUUGGUCCCAAACCAAGUACUCUACCAAGCUGAGCUACUUCCCGAGUUAAAUAGAAAAAUGCACCCUAGAGGAGUCGAACCUCUAACCGC ((((((((.((((.......))))......((((((.....))))))..(((.........)))...)))))))).....................((((((.......))))))..... ( -31.20) >consensus UCGGGAAGACAGGAUUCGAACCUGCGACCCCUUGGUCCCAAACCAAGUGCUCUACCAAGCUGAGCUACUUCCCGUA_______AAAAAAUGCGCCCGAUAGGAGUCGAACCUAUAACCUC .(((((((.((((.......))))......((((((.....)))))).((((.........))))..))))))).......................(((((.......)))))...... (-27.18 = -27.50 + 0.32)

| Location | 1,976,242 – 1,976,358 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.53 |
| Mean single sequence MFE | -30.68 |
| Consensus MFE | -27.42 |
| Energy contribution | -26.14 |
| Covariance contribution | -1.28 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1976242 116 + 2809422/80-200 ---AAUUAACGCGCCCGAUAGGAGUCGAACCCAUAACCUCUUGAUCCGUAGUCAAACGCUCUAUCCAAUUGAGCUACGGGCGCAUAUGUUUUUAUUGAAA-AUGGUGCCGAGGACCGGAA ---.......(((((((...(((.((((............)))))))..........((((.........))))..))))))).................-.((((.(....)))))... ( -29.40) >sy_ha.0 1576076 108 - 2685015/80-200 ---GAG-AACGCGCCCGAUAGGAGUCGAACCCAUAACCUCUUGAUCCGUAGUCAAACGCUCUAUCCAAUUGAGCUACGGGCGCAAA-------AUUAUAU-AUGGUGCCGAGGACCGGAA ---...-...(((((((...(((.((((............)))))))..........((((.........))))..)))))))...-------.......-.((((.(....)))))... ( -29.40) >sy_sa.0 1551116 113 - 2516575/80-200 ---AAUUAGUGCGCCCAAUAGGGGUCGAACCUAUAACCUCUUGAUUCGUAGUCAAGUACUCUAUCCAGUUGAGCUAUGGGCGCAUAA----UUUUUAUAGGAUGAUGCCGAGGACCGGAA ---.....((((((((((((((.......))))).....((((((.....))))))..(((.........)))...)))))))))..----................(((.....))).. ( -30.80) >sp_ag.0 2132345 105 - 2160267/80-200 ----AAAUAUGCACCCUAGAGGAGUCGAACCUCUAACCGCCUGAUUCGUAGUCAGGUACUCUAUCCAGUUGAGCUAAGGGUGCUAAA-----UAUUAUA------UGCCGAGGACCGGAA ----......((((((((((((.......))))))...(((((((.....))))))).(((.........)))....))))))....-----.......------..(((.....))).. ( -31.90) >sp_pn.0 1691504 107 + 2038615/80-200 AUAGAAAAAUGCACCCUAGAGGAGUCGAACCUCUAACCGCCUGAUUCGUAGUCAGGUACUCUAUCCAGUUGAGCUAAGGGUGCUCCA-----UAUUA--------UGCCGAGGACCGGAA ..........((((((((((((.......))))))...(((((((.....))))))).(((.........)))....))))))....-----.....--------..(((.....))).. ( -31.90) >consensus ___AAAAAAUGCGCCCGAUAGGAGUCGAACCUAUAACCUCUUGAUUCGUAGUCAAGUACUCUAUCCAGUUGAGCUAAGGGCGCAAAA_____UAUUAUA__AUG_UGCCGAGGACCGGAA ..........((((((((((((.......))))).....((((((.....))))))..(((.........)))...)))))))........................(((.....))).. (-27.42 = -26.14 + -1.28)

| Location | 1,976,242 – 1,976,358 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.53 |
| Mean single sequence MFE | -30.14 |
| Consensus MFE | -29.58 |
| Energy contribution | -26.78 |
| Covariance contribution | -2.80 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.71 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842628 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1976242 116 + 2809422/80-200 UUCCGGUCCUCGGCACCAU-UUUCAAUAAAAACAUAUGCGCCCGUAGCUCAAUUGGAUAGAGCGUUUGACUACGGAUCAAGAGGUUAUGGGUUCGACUCCUAUCGGGCGCGUUAAUU--- ....((((....).)))..-...............(((((((((..((((.........))))(((.((((...((((....))))...)))).)))......))))))))).....--- ( -33.00) >sy_ha.0 1576076 108 - 2685015/80-200 UUCCGGUCCUCGGCACCAU-AUAUAAU-------UUUGCGCCCGUAGCUCAAUUGGAUAGAGCGUUUGACUACGGAUCAAGAGGUUAUGGGUUCGACUCCUAUCGGGCGCGUU-CUC--- ....((((....).)))..-.......-------...(((((((..((((.........))))(((.((((...((((....))))...)))).)))......)))))))...-...--- ( -32.10) >sy_sa.0 1551116 113 - 2516575/80-200 UUCCGGUCCUCGGCAUCAUCCUAUAAAAA----UUAUGCGCCCAUAGCUCAACUGGAUAGAGUACUUGACUACGAAUCAAGAGGUUAUAGGUUCGACCCCUAUUGGGCGCACUAAUU--- ..(((.....)))..............((----(((((((((((..((((.........)))).(((((.......))))).....(((((.......))))))))))))).)))))--- ( -27.00) >sp_ag.0 2132345 105 - 2160267/80-200 UUCCGGUCCUCGGCA------UAUAAUA-----UUUAGCACCCUUAGCUCAACUGGAUAGAGUACCUGACUACGAAUCAGGCGGUUAGAGGUUCGACUCCUCUAGGGUGCAUAUUU---- ..(((.....)))..------.......-----....((((((...((((.........)))).(((((.......)))))....((((((.......))))))))))))......---- ( -29.30) >sp_pn.0 1691504 107 + 2038615/80-200 UUCCGGUCCUCGGCA--------UAAUA-----UGGAGCACCCUUAGCUCAACUGGAUAGAGUACCUGACUACGAAUCAGGCGGUUAGAGGUUCGACUCCUCUAGGGUGCAUUUUUCUAU ..(((.....)))..--------.....-----....((((((...((((.........)))).(((((.......)))))....((((((.......)))))))))))).......... ( -29.30) >consensus UUCCGGUCCUCGGCA_CAU__UAUAAUA_____UUAUGCGCCCGUAGCUCAACUGGAUAGAGUACUUGACUACGAAUCAAGAGGUUAUAGGUUCGACUCCUAUAGGGCGCAUUAAUC___ ..(((.....)))........................(((((((..((((.........)))).(((((.......))))).....(((((.......)))))))))))).......... (-29.58 = -26.78 + -2.80) # Strand winner: reverse (0.68)

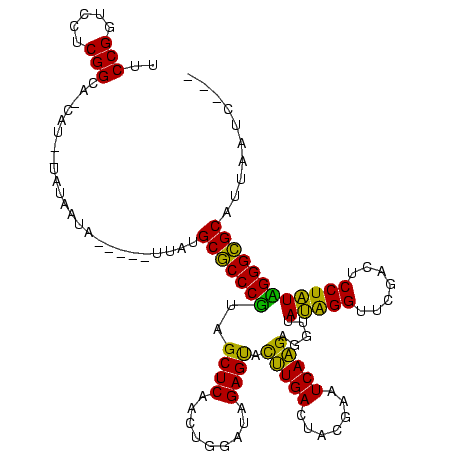
| Location | 1,976,242 – 1,976,358 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.77 |
| Mean single sequence MFE | -37.78 |
| Consensus MFE | -32.22 |
| Energy contribution | -32.14 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1976242 116 + 2809422/160-280 CGCAUAUGUUUUUAUUGAAA-AUGGUGCCGAGGACCGGAAUCGAACCGGUACGGUGAUCACUCACCGCAGGAUUUUAAGUCCUGUGCGUCUGCCAGUUCCGCCACCCCGGCACUAUA--- ....................-(((((((((.((...((....((((.((((.(((((....)))))(((((((.....))))))).....)))).))))..))..))))))))))).--- ( -43.40) >sy_ha.0 1576076 112 - 2685015/160-280 CGCAAA-------AUUAUAU-AUGGUGCCGAGGACCGGAAUCGAACCGGUACGGUGAUCUCUCACCGCAGGAUUUUAAGUCCUGUGCGUCUGCCAGUUCCGCCACCCCGGCACUAUAUUA ......-------.....((-(((((((((.((...((....((((.((((.(((((....)))))(((((((.....))))))).....)))).))))..))..))))))))))))).. ( -45.10) >sy_sa.0 1551116 113 - 2516575/160-280 CGCAUAA----UUUUUAUAGGAUGAUGCCGAGGACCGGAAUCGAACCGGUACGAUAAUCACUUAUCGCAGGAUUUUAAGUCCUGUGCGUCUGCCAGUUCCGCCACCCCGGCAAAAUA--- .......----..............(((((.((...((....((((.((((.(((((....)))))(((((((.....))))))).....)))).))))..))..))))))).....--- ( -32.80) >sp_ag.0 2132345 105 - 2160267/160-280 UGCUAAA-----UAUUAUA------UGCCGAGGACCGGAAUCGAACCGGUACGAUGUUUACCA-UCGCAGGAUUUUAAGUCCUGUGCGUCUGCCAGUUCCGCCACCCCGGCCUCUAA--- .......-----.......------....((((.((((....((((.((((.(((((......-..(((((((.....)))))))))))))))).)))).......))))))))...--- ( -34.80) >sp_pn.0 1691504 103 + 2038615/160-280 UGCUCCA-----UAUUA--------UGCCGAGGACCGGAAUCGAACCGGUACGAUCGUUACCAAUCGCAGGAUUUUAAGUCCUGUGCGUCUGCCAGUUCCGCCACCCCGGCCUCUC---- .......-----.....--------....((((.((((....((((.((((.((.(((........(((((((.....)))))))))))))))).)))).......))))))))..---- ( -32.80) >consensus CGCAAAA_____UAUUAUA__AUG_UGCCGAGGACCGGAAUCGAACCGGUACGAUGAUCACUAAUCGCAGGAUUUUAAGUCCUGUGCGUCUGCCAGUUCCGCCACCCCGGCACCAUA___ ..........................((((.((...((....((((.((((.(((((....)))))(((((((.....))))))).....)))).))))..))..))))))......... (-32.22 = -32.14 + -0.08)


| Location | 1,976,242 – 1,976,358 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.77 |
| Mean single sequence MFE | -39.78 |
| Consensus MFE | -36.52 |
| Energy contribution | -36.00 |
| Covariance contribution | -0.52 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1976242 116 + 2809422/160-280 ---UAUAGUGCCGGGGUGGCGGAACUGGCAGACGCACAGGACUUAAAAUCCUGCGGUGAGUGAUCACCGUACCGGUUCGAUUCCGGUCCUCGGCACCAU-UUUCAAUAAAAACAUAUGCG ---....(((((((((.(.(((((((((........(((((.......)))))((((((....))))))..))))).....)))).))))))))))...-.................... ( -43.60) >sy_ha.0 1576076 112 - 2685015/160-280 UAAUAUAGUGCCGGGGUGGCGGAACUGGCAGACGCACAGGACUUAAAAUCCUGCGGUGAGAGAUCACCGUACCGGUUCGAUUCCGGUCCUCGGCACCAU-AUAUAAU-------UUUGCG ..((((.(((((((((.(.(((((((((........(((((.......)))))((((((....))))))..))))).....)))).)))))))))).))-)).....-------...... ( -44.00) >sy_sa.0 1551116 113 - 2516575/160-280 ---UAUUUUGCCGGGGUGGCGGAACUGGCAGACGCACAGGACUUAAAAUCCUGCGAUAAGUGAUUAUCGUACCGGUUCGAUUCCGGUCCUCGGCAUCAUCCUAUAAAAA----UUAUGCG ---.....((((((((.(.(((((((((........(((((.......)))))((((((....))))))..))))).....)))).)))))))))..............----....... ( -36.40) >sp_ag.0 2132345 105 - 2160267/160-280 ---UUAGAGGCCGGGGUGGCGGAACUGGCAGACGCACAGGACUUAAAAUCCUGCGA-UGGUAAACAUCGUACCGGUUCGAUUCCGGUCCUCGGCA------UAUAAUA-----UUUAGCA ---...(((((((((((....(((((((........(((((.......)))))(((-((.....)))))..))))))).))))))).))))....------.......-----....... ( -36.90) >sp_pn.0 1691504 103 + 2038615/160-280 ----GAGAGGCCGGGGUGGCGGAACUGGCAGACGCACAGGACUUAAAAUCCUGCGAUUGGUAACGAUCGUACCGGUUCGAUUCCGGUCCUCGGCA--------UAAUA-----UGGAGCA ----(((.(((((((((....(((((((........(((((.......)))))((((((....))))))..))))))).)))))))))))).((.--------.....-----....)). ( -38.00) >consensus ___UAUAAUGCCGGGGUGGCGGAACUGGCAGACGCACAGGACUUAAAAUCCUGCGAUGAGUGAUCAUCGUACCGGUUCGAUUCCGGUCCUCGGCA_CAU__UAUAAUA_____UUAUGCG .........(((((((.(.(((((((((........(((((.......)))))((((((....))))))..))))).....)))).)))))))).......................... (-36.52 = -36.00 + -0.52) # Strand winner: reverse (0.99)

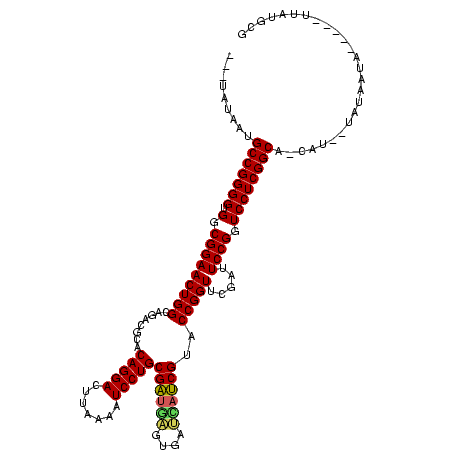

| Location | 1,976,242 – 1,976,344 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.91 |
| Mean single sequence MFE | -36.88 |
| Consensus MFE | -34.54 |
| Energy contribution | -33.26 |
| Covariance contribution | -1.28 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.837077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1976242 102 + 2809422/480-600 -----AUGCGGGUGAAGGGAGUCGAACCCCCACGCCGUAAGGCGCUAGAUCCUAAGUCUAGUGCGUCUGCCAAUUCCGCCACACCCGCAA----------AUG---GUGAGCCAUAGAGG -----.((((((((..(((.(.....).)))..(((....)))(((((((.....)))))))(((...........)))..)))))))).----------(((---(....))))..... ( -38.90) >sy_ha.0 1576076 105 - 2685015/480-600 -----AUGCGGGUGAAGGGAGUCGAACCCCCACGCCGUAAGGCGCUAGAUCCUAAGUCUAGUGCGUCUGCCAAUUCCGCCACACCCGCGU----------AUACAUGUGAGCCAUAGAGG -----(((((((((..(((.(.....).)))..(((....)))(((((((.....)))))))(((...........)))..)))))))))----------.................... ( -35.60) >sy_sa.0 1551116 115 - 2516575/480-600 -----AUGCGGGUGAAGGGAGUCGAACCCCCACGCCGUAAGGCGCUAGAUCCUAAGUCUAGUGCGUCUGCCAAUUCCGCCACACCCGCGAUAUUAAAAUGAUAAUGGUGAGCCAUAGUGG -----.((((((((..(((.(.....).)))..(((....)))(((((((.....)))))))(((...........)))..))))))))..............((((....))))..... ( -38.20) >sp_ag.0 2132345 110 - 2160267/480-600 UACUAAUGCGGGUGAAGGGACUUGAACCCCCACGCCGUUAAGCGCCAGAUCCUAAAUCUGGUGCGUCUGCCAAUUCCGCCACACCCGCAU----------UUCUAAAUGACCCGUACUGG ((..((((((((((..(((.........)))..((.(....(((((((((.....)))))))))..).))...........)))))))))----------)..))............... ( -37.20) >sp_pn.0 1691504 106 + 2038615/480-600 UCUUAAUGCGGGUUAAGGGACUUGAACCCCCACGCCGUUAAGCGCCAGAUCCUAAAUCUGGUGCGUCUGCCAAUUCCGCCAAACCCGCAU----------AU----AUGACCCGUACUGG .....(((((((((..(((.........)))..((.(....(((((((((.....)))))))))..).))...........)))))))))----------..----.............. ( -34.50) >consensus _____AUGCGGGUGAAGGGAGUCGAACCCCCACGCCGUAAGGCGCUAGAUCCUAAGUCUAGUGCGUCUGCCAAUUCCGCCACACCCGCAU__________AUA___GUGAGCCAUAGUGG ......((((((((..(((.........)))..((.(....(((((((((.....)))))))))..).))...........))))))))............................... (-34.54 = -33.26 + -1.28)

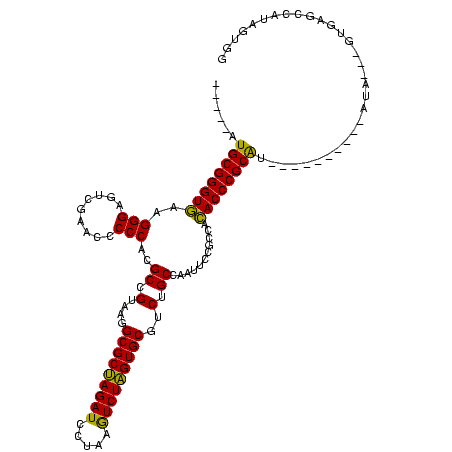
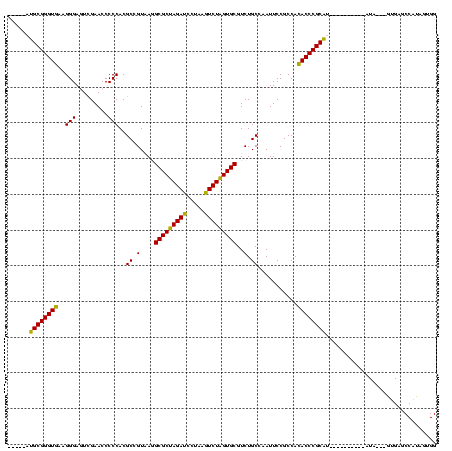
| Location | 1,976,242 – 1,976,349 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.67 |
| Mean single sequence MFE | -31.92 |
| Consensus MFE | -31.20 |
| Energy contribution | -28.96 |
| Covariance contribution | -2.24 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1976242 107 + 2809422/520-640 GGCGCUAGAUCCUAAGUCUAGUGCGUCUGCCAAUUCCGCCACACCCGCAA----------AUG---GUGAGCCAUAGAGGAUUCGAACCUCUGACCCUCUGAUUAAAAGUCAGAUGCUCU .(((((((((.....)))))))))....((((.((.((.......)).))----------.))---))((((..((((((.......))))))....((((((.....)))))).)))). ( -34.10) >sy_ha.0 1576076 110 - 2685015/520-640 GGCGCUAGAUCCUAAGUCUAGUGCGUCUGCCAAUUCCGCCACACCCGCGU----------AUACAUGUGAGCCAUAGAGGAUUCGAACCUCUGACCCUCUGAUUAAAAGUCAGAUGCUCU .(((((((((.....)))))))))............(((.......))).----------........((((..((((((.......))))))....((((((.....)))))).)))). ( -32.20) >sy_sa.0 1551116 120 - 2516575/520-640 GGCGCUAGAUCCUAAGUCUAGUGCGUCUGCCAAUUCCGCCACACCCGCGAUAUUAAAAUGAUAAUGGUGAGCCAUAGUGGGCUCGAACCACUGACCCUCUGAUUAAAAGUCAGAUGCUCU .(((((((((.....)))))))))....((((....(((.......))).((((.....)))).))))((((..((((((.......))))))....((((((.....)))))).)))). ( -37.30) >sp_ag.0 2132345 110 - 2160267/520-640 AGCGCCAGAUCCUAAAUCUGGUGCGUCUGCCAAUUCCGCCACACCCGCAU----------UUCUAAAUGACCCGUACUGGGCUCGAACCAGUGACCCAUUGAUUAAAAGUCAAUUGCUCU .(((((((((.....)))))))))......................(((.----------..............((((((.......))))))....((((((.....)))))))))... ( -28.00) >sp_pn.0 1691504 106 + 2038615/520-640 AGCGCCAGAUCCUAAAUCUGGUGCGUCUGCCAAUUCCGCCAAACCCGCAU----------AU----AUGACCCGUACUGGGCUCGAACCAGUGACCCAUUGAUUAAAAGUCAAUUGCUCU .(((((((((.....)))))))))......................(((.----------..----........((((((.......))))))....((((((.....)))))))))... ( -28.00) >consensus GGCGCUAGAUCCUAAGUCUAGUGCGUCUGCCAAUUCCGCCACACCCGCAU__________AUA___GUGAGCCAUAGUGGGCUCGAACCACUGACCCUCUGAUUAAAAGUCAGAUGCUCU .(((((((((.....))))))))).............((.......))....................((((..((((((.......))))))....((((((.....)))))).)))). (-31.20 = -28.96 + -2.24)

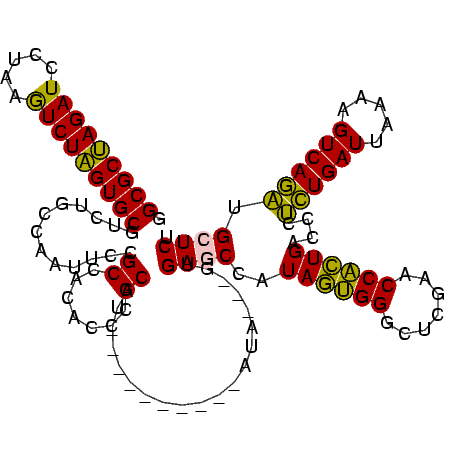
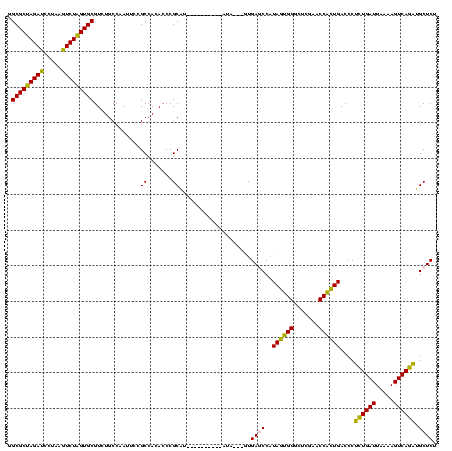
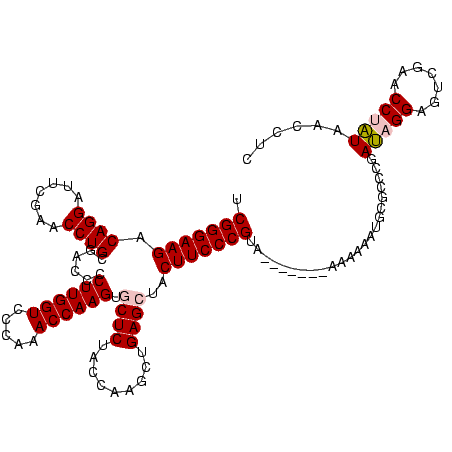
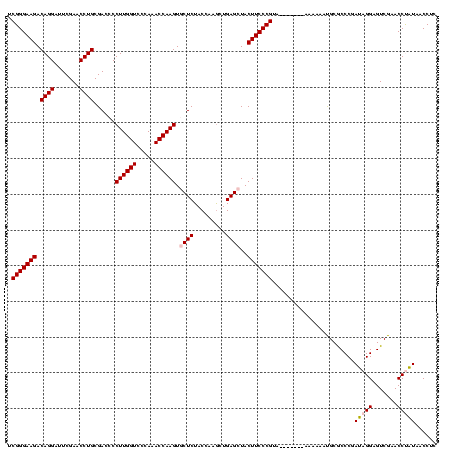
| Location | 1,976,242 – 1,976,346 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.91 |
| Mean single sequence MFE | -31.53 |
| Consensus MFE | -28.46 |
| Energy contribution | -25.58 |
| Covariance contribution | -2.88 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.998549 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1976242 104 + 2809422/560-680 ACACCCGCAA----------AUG---GUGAGCCAUAGAGGAUUCGAACCUCUGACCCUCUGAUUAAAAGUCAGAUGCUCUACCAACUGAGCUAAUGGCUCU---UCCAUGGUGCCGGCCA ...((.(((.----------(((---(.((((((((((((.......))))).....((((((.....)))))).((((........))))..))))))).---.))))..))).))... ( -39.20) >sy_ha.0 1576076 110 - 2685015/560-680 ACACCCGCGU----------AUACAUGUGAGCCAUAGAGGAUUCGAACCUCUGACCCUCUGAUUAAAAGUCAGAUGCUCUACCAACUGAGCUAAUGGCUCAAAAUAGAUGGUGCCGGCCA ....(((.((----------((.....(((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))).........)))))))... ( -35.34) >sy_sa.0 1551116 119 - 2516575/560-680 ACACCCGCGAUAUUAAAAUGAUAAUGGUGAGCCAUAGUGGGCUCGAACCACUGACCCUCUGAUUAAAAGUCAGAUGCUCUACCAACUGAGCUAAUGGCUCAUC-AAAAUGGUGCCGGCCA .((((.....((((.....)))).((((((((((((((((.......))))).....((((((.....)))))).((((........))))..))))))))))-)....))))....... ( -39.20) >sp_ag.0 2132345 92 - 2160267/560-680 ACACCCGCAU----------UUCUAAAUGACCCGUACUGGGCUCGAACCAGUGACCCAUUGAUUAAAAGUCAAUUGCUCUACCAACUGAGCUAACGAGUCU---A--------------- ..........----------........(((.((((((((.......))))).....((((((.....)))))).((((........))))..))).))).---.--------------- ( -20.50) >sp_pn.0 1691504 103 + 2038615/560-680 AAACCCGCAU----------AU----AUGACCCGUACUGGGCUCGAACCAGUGACCCAUUGAUUAAAAGUCAAUUGCUCUACCAACUGAGCUAACGAGUCU---AAAAUAACUUGCGUUA .....((((.----------.(----(((((.((((((((.......))))).....((((((.....)))))).((((........))))..))).))).---...)))...))))... ( -23.40) >consensus ACACCCGCAU__________AUA___GUGAGCCAUAGUGGGCUCGAACCACUGACCCUCUGAUUAAAAGUCAGAUGCUCUACCAACUGAGCUAAUGGCUCU___AAAAUGGUGCCGGCCA ............................((((((((((((.......))))).....((((((.....)))))).((((........))))..)))))))........((((....)))) (-28.46 = -25.58 + -2.88)

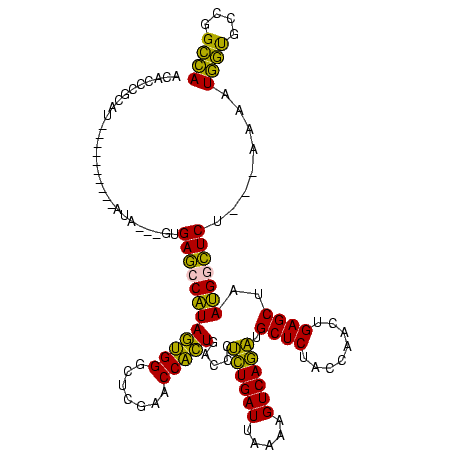
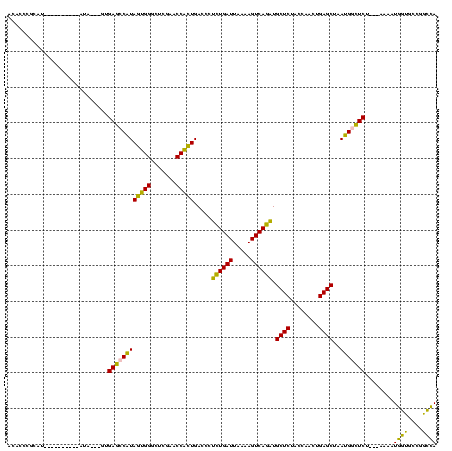
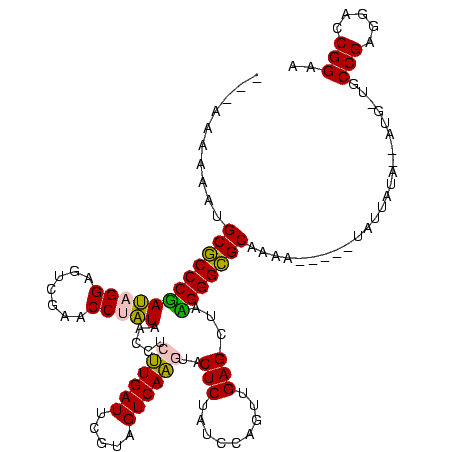
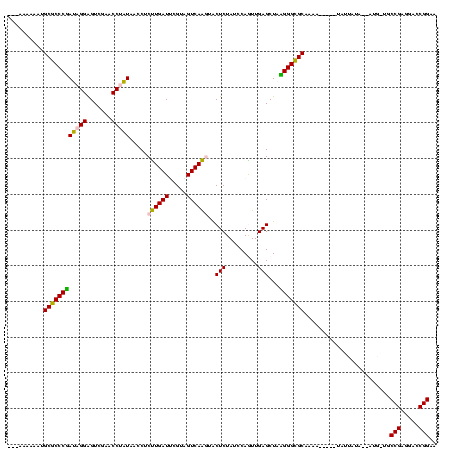
| Location | 1,976,242 – 1,976,346 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.91 |
| Mean single sequence MFE | -34.60 |
| Consensus MFE | -28.60 |
| Energy contribution | -26.60 |
| Covariance contribution | -2.00 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.997483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1976242 104 + 2809422/560-680 UGGCCGGCACCAUGGA---AGAGCCAUUAGCUCAGUUGGUAGAGCAUCUGACUUUUAAUCAGAGGGUCAGAGGUUCGAAUCCUCUAUGGCUCAC---CAU----------UUGCGGGUGU ..(((.(((..((((.---.(((((((......((..((..((((.((((((((((.....)))))))))).))))....)).))))))))).)---)))----------.))).))).. ( -40.80) >sy_ha.0 1576076 110 - 2685015/560-680 UGGCCGGCACCAUCUAUUUUGAGCCAUUAGCUCAGUUGGUAGAGCAUCUGACUUUUAAUCAGAGGGUCAGAGGUUCGAAUCCUCUAUGGCUCACAUGUAU----------ACGCGGGUGU ..(((.((.........((((((((....((((........)))).((((((((((.....)))))))))))))))))).....((((.....))))...----------..)).))).. ( -36.70) >sy_sa.0 1551116 119 - 2516575/560-680 UGGCCGGCACCAUUUU-GAUGAGCCAUUAGCUCAGUUGGUAGAGCAUCUGACUUUUAAUCAGAGGGUCAGUGGUUCGAGCCCACUAUGGCUCACCAUUAUCAUUUUAAUAUCGCGGGUGU ..(((.((........-..(((((.....)))))..((((.((((..(((((((((.....)))))))))..))))(((((......)))))))))................)).))).. ( -41.70) >sp_ag.0 2132345 92 - 2160267/560-680 ---------------U---AGACUCGUUAGCUCAGUUGGUAGAGCAAUUGACUUUUAAUCAAUGGGUCACUGGUUCGAGCCCAGUACGGGUCAUUUAGAA----------AUGCGGGUGU ---------------.---.(((((((..((((((..(((.((.(.(((((.......))))).).)))))..)).)))).....)))))))........----------.......... ( -24.60) >sp_pn.0 1691504 103 + 2038615/560-680 UAACGCAAGUUAUUUU---AGACUCGUUAGCUCAGUUGGUAGAGCAAUUGACUUUUAAUCAAUGGGUCACUGGUUCGAGCCCAGUACGGGUCAU----AU----------AUGCGGGUUU ((((....))))....---((((((((..((((........))))...(((((..((.....((((((........)).)))).))..))))).----..----------..)))))))) ( -29.20) >consensus UGGCCGGCACCAUUUU___AGAGCCAUUAGCUCAGUUGGUAGAGCAUCUGACUUUUAAUCAGAGGGUCAGUGGUUCGAGCCCACUAUGGCUCAC___UAU__________AUGCGGGUGU ....................(((((((..((((........))))....(((((((.....)))))))(((((.......))))))))))))............................ (-28.60 = -26.60 + -2.00) # Strand winner: reverse (0.98)

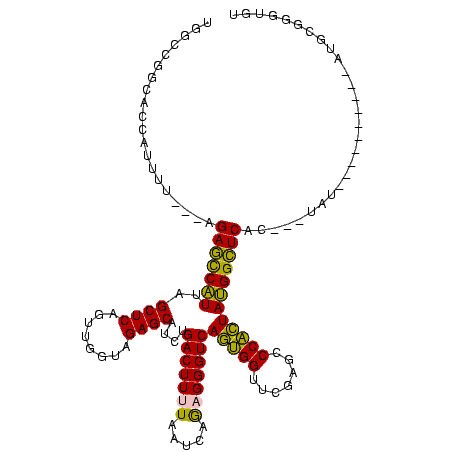
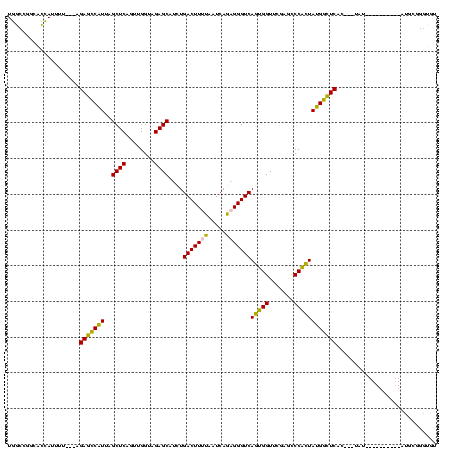
| Location | 1,976,242 – 1,976,359 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.94 |
| Mean single sequence MFE | -30.26 |
| Consensus MFE | -14.59 |
| Energy contribution | -13.15 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.48 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.529650 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1976242 117 + 2809422/600-720 AUUCGAACCUCUGACCCUCUGAUUAAAAGUCAGAUGCUCUACCAACUGAGCUAAUGGCUCU---UCCAUGGUGCCGGCCAGAGGACUUGAACCCCCAACCUACUGAUUACAAGUCAGUUG .(((((.((((((.((.((((((.....)))))).((((........))))....(((.((---.....)).))))).))))))..)))))..........((((((.....)))))).. ( -36.40) >sy_ha.0 1576076 120 - 2685015/600-720 AUUCGAACCUCUGACCCUCUGAUUAAAAGUCAGAUGCUCUACCAACUGAGCUAAUGGCUCAAAAUAGAUGGUGCCGGCCAGAGGACUUGAACCCCCAACCUACUGAUUACAAGUCAGUUG .(((((.((((((.((.((((((.....)))))).((((........))))....((((((.......))).))))).))))))..)))))..........((((((.....)))))).. ( -35.10) >sy_sa.0 1551116 119 - 2516575/600-720 GCUCGAACCACUGACCCUCUGAUUAAAAGUCAGAUGCUCUACCAACUGAGCUAAUGGCUCAUC-AAAAUGGUGCCGGCCAGAGGACUUGAACCCCCAACCUACUGAUUACAAGUCAGUUG ..((((.((.(((.((.((((((.....)))))).((((........))))....(((((((.-...)))).))))).))).))..))))...........((((((.....)))))).. ( -30.70) >sp_ag.0 2132345 95 - 2160267/600-720 GCUCGAACCAGUGACCCAUUGAUUAAAAGUCAAUUGCUCUACCAACUGAGCUAACGAGUCU---A----------------------CGGUCCCGACGGGAAUCGAACCCGCGAUCUUCG ...((((((.(((((.(((((((.....)))))).((((........))))....).))).---)----------------------))))..((.((((.......))))))....))) ( -22.90) >sp_pn.0 1691504 117 + 2038615/600-720 GCUCGAACCAGUGACCCAUUGAUUAAAAGUCAAUUGCUCUACCAACUGAGCUAACGAGUCU---AAAAUAACUUGCGUUAUCUUAAACGGUCCCGACGGGAAUCGAACCCGCGAUCUUCG ...((((.....((((.((((((.....)))))).((((........))))......((.(---((.(((((....))))).))).)))))).((.((((.......))))))...)))) ( -26.20) >consensus GCUCGAACCACUGACCCUCUGAUUAAAAGUCAGAUGCUCUACCAACUGAGCUAAUGGCUCU___AAAAUGGUGCCGGCCAGAGGACUUGAACCCCCAACCUACUGAUUACAAGUCAGUUG .................((((((.....)))))).((((........))))...((((..........((((....))))...((((((....................)))))).)))) (-14.59 = -13.15 + -1.44)

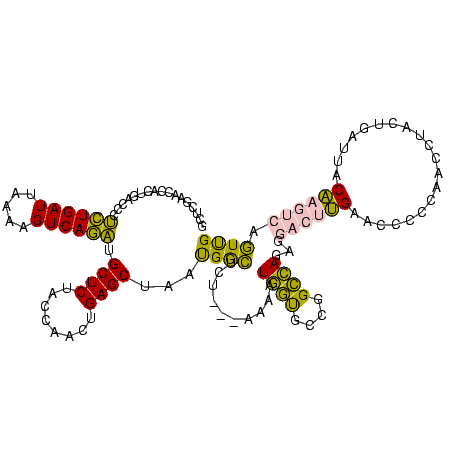
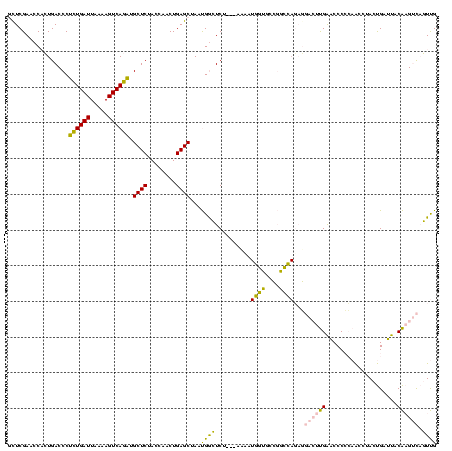
| Location | 1,976,242 – 1,976,359 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.94 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -24.80 |
| Energy contribution | -25.12 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.62 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1976242 117 + 2809422/600-720 CAACUGACUUGUAAUCAGUAGGUUGGGGGUUCAAGUCCUCUGGCCGGCACCAUGGA---AGAGCCAUUAGCUCAGUUGGUAGAGCAUCUGACUUUUAAUCAGAGGGUCAGAGGUUCGAAU (((((((((.((........(((..((((.......))))..)))(((........---...))))).)).)))))))...((((.((((((((((.....)))))))))).)))).... ( -43.40) >sy_ha.0 1576076 120 - 2685015/600-720 CAACUGACUUGUAAUCAGUAGGUUGGGGGUUCAAGUCCUCUGGCCGGCACCAUCUAUUUUGAGCCAUUAGCUCAGUUGGUAGAGCAUCUGACUUUUAAUCAGAGGGUCAGAGGUUCGAAU ..(((((.......))))).(((..((((.......))))..)))............((((((((....((((........)))).((((((((((.....)))))))))))))))))). ( -44.10) >sy_sa.0 1551116 119 - 2516575/600-720 CAACUGACUUGUAAUCAGUAGGUUGGGGGUUCAAGUCCUCUGGCCGGCACCAUUUU-GAUGAGCCAUUAGCUCAGUUGGUAGAGCAUCUGACUUUUAAUCAGAGGGUCAGUGGUUCGAGC (((((((((.((........(((..((((.......))))..)))(((..(((...-.))).))))).)).)))))))...((((..(((((((((.....)))))))))..)))).... ( -42.20) >sp_ag.0 2132345 95 - 2160267/600-720 CGAAGAUCGCGGGUUCGAUUCCCGUCGGGACCG----------------------U---AGACUCGUUAGCUCAGUUGGUAGAGCAAUUGACUUUUAAUCAAUGGGUCACUGGUUCGAGC .......((((((.......)))).))((((((----------------------.---.((((((((((.(((((((......))))))))).......))))))))..)))))).... ( -29.41) >sp_pn.0 1691504 117 + 2038615/600-720 CGAAGAUCGCGGGUUCGAUUCCCGUCGGGACCGUUUAAGAUAACGCAAGUUAUUUU---AGACUCGUUAGCUCAGUUGGUAGAGCAAUUGACUUUUAAUCAAUGGGUCACUGGUUCGAGC ((((....(((((.......)))))(((((((((((((((((((....))))))))---))))......((((........)))).(((((.......))))).)))).))).))))... ( -40.40) >consensus CAACUGACUUGUAAUCAGUAGGUUGGGGGUUCAAGUCCUCUGGCCGGCACCAUUUU___AGAGCCAUUAGCUCAGUUGGUAGAGCAUCUGACUUUUAAUCAGAGGGUCAGUGGUUCGAGC .....((((((.((((...........)))))))))).......................(((((....((((........)))).((((((((((.....))))))))))))))).... (-24.80 = -25.12 + 0.32) # Strand winner: reverse (1.00)

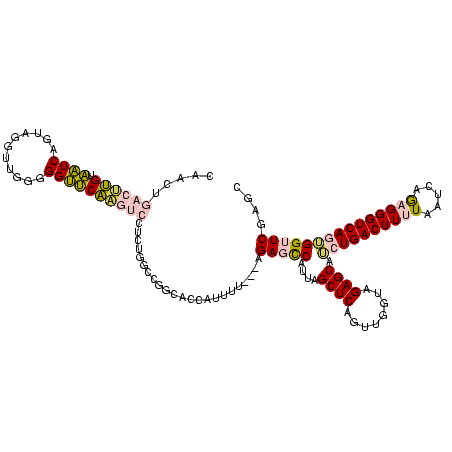
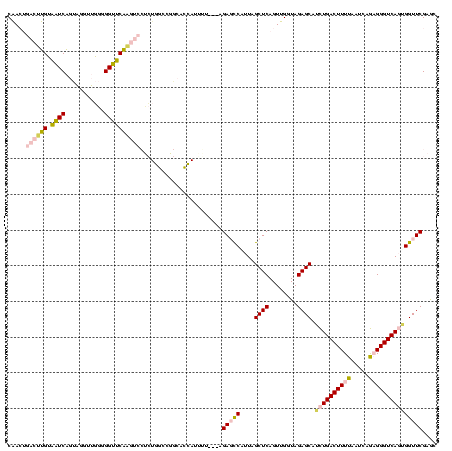
| Location | 1,976,242 – 1,976,355 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 56.85 |
| Mean single sequence MFE | -33.56 |
| Consensus MFE | -14.67 |
| Energy contribution | -17.99 |
| Covariance contribution | 3.32 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1976242 113 + 2809422/640-760 ACCAACUGAGCUAAUGGCUCU---UCCAUGGUGCCGGCCAGAGGACUUGAACCCCCAACCUACUGAUUACAAGUCAGUUGCUCUACCAAUUGAGCUAGGCCGGCAAUAUGUAA----GAA .......((((.....)))).---..((((.((((((((..(((..(((......))))))((((((.....)))))).((((........))))..)))))))).))))...----... ( -39.10) >sy_ha.0 1576076 120 - 2685015/640-760 ACCAACUGAGCUAAUGGCUCAAAAUAGAUGGUGCCGGCCAGAGGACUUGAACCCCCAACCUACUGAUUACAAGUCAGUUGCUCUACCAAUUGAGCUAGGCCGGCAAUAUAUUGUUUUGAU .......((((.....))))((((((((((.((((((((..(((..(((......))))))((((((.....)))))).((((........))))..)))))))).))).)))))))... ( -40.30) >sy_sa.0 1551116 112 - 2516575/640-760 ACCAACUGAGCUAAUGGCUCAUC-AAAAUGGUGCCGGCCAGAGGACUUGAACCCCCAACCUACUGAUUACAAGUCAGUUGCUCUACCAGUUGAGCUAGGCCGGC--UAAAUUA-----AA ((((..(((((.....)))))..-....))))(((((((..(((..(((......))))))((((((.....)))))).((((........))))..)))))))--.......-----.. ( -37.80) >sp_ag.0 2132345 90 - 2160267/640-760 ACCAACUGAGCUAACGAGUCU---A----------------------CGGUCCCGACGGGAAUCGAACCCGCGAUCUUCGCCGUGACAGGGCGACGUGAUAACCGCUACACUA-----CG ..............(((((.(---(----------------------((((..((.((((.......))))))(((.(((((.......)))))...))).)))).)).))).-----)) ( -23.20) >sp_pn.0 1691504 112 + 2038615/640-760 ACCAACUGAGCUAACGAGUCU---AAAAUAACUUGCGUUAUCUUAAACGGUCCCGACGGGAAUCGAACCCGCGAUCUUCGCCGUGACAGGGCGACGUGAUAACCGCUACACUA-----CG ......(((((...(((((..---......))))).((((((....(((....((.((((.......))))))....(((((.......)))))))))))))).))).))...-----.. ( -27.40) >consensus ACCAACUGAGCUAAUGGCUCU___AAAAUGGUGCCGGCCAGAGGACUUGAACCCCCAACCUACUGAUUACAAGUCAGUUGCUCUACCAGUUGAGCUAGGCCGGCAAUACAUUA_____AA .......((((.....))))...........((((((((....((((((....................))))))....((((........))))..))))))))............... (-14.67 = -17.99 + 3.32)

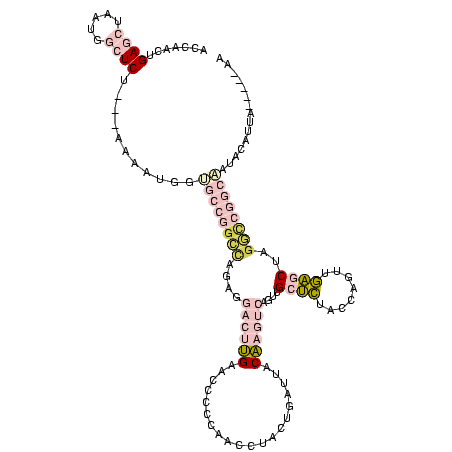
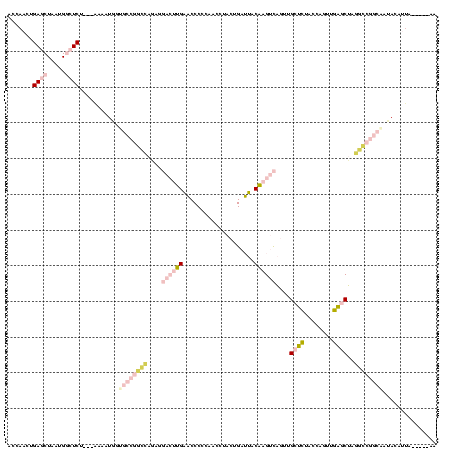
| Location | 1,976,242 – 1,976,355 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 56.85 |
| Mean single sequence MFE | -37.18 |
| Consensus MFE | -20.86 |
| Energy contribution | -21.46 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1976242 113 + 2809422/640-760 UUC----UUACAUAUUGCCGGCCUAGCUCAAUUGGUAGAGCAACUGACUUGUAAUCAGUAGGUUGGGGGUUCAAGUCCUCUGGCCGGCACCAUGGA---AGAGCCAUUAGCUCAGUUGGU .((----((.(((..((((((((..((((........)))).(((((.......))))).....(((((.......)))))))))))))..))).)---)))((((..........)))) ( -39.40) >sy_ha.0 1576076 120 - 2685015/640-760 AUCAAAACAAUAUAUUGCCGGCCUAGCUCAAUUGGUAGAGCAACUGACUUGUAAUCAGUAGGUUGGGGGUUCAAGUCCUCUGGCCGGCACCAUCUAUUUUGAGCCAUUAGCUCAGUUGGU ................(((((((..((((........)))).(((((.......))))).....(((((.......))))))))))))((((......((((((.....)))))).)))) ( -39.90) >sy_sa.0 1551116 112 - 2516575/640-760 UU-----UAAUUUA--GCCGGCCUAGCUCAACUGGUAGAGCAACUGACUUGUAAUCAGUAGGUUGGGGGUUCAAGUCCUCUGGCCGGCACCAUUUU-GAUGAGCCAUUAGCUCAGUUGGU ..-----.......--(((((((..((((........)))).(((((.......))))).....(((((.......))))))))))))((((....-..(((((.....)))))..)))) ( -40.30) >sp_ag.0 2132345 90 - 2160267/640-760 CG-----UAGUGUAGCGGUUAUCACGUCGCCCUGUCACGGCGAAGAUCGCGGGUUCGAUUCCCGUCGGGACCG----------------------U---AGACUCGUUAGCUCAGUUGGU ((-----.(((.((.(((((......(((((.......)))))...(((((((.......)))).))))))))----------------------)---).))))).............. ( -26.90) >sp_pn.0 1691504 112 + 2038615/640-760 CG-----UAGUGUAGCGGUUAUCACGUCGCCCUGUCACGGCGAAGAUCGCGGGUUCGAUUCCCGUCGGGACCGUUUAAGAUAACGCAAGUUAUUUU---AGACUCGUUAGCUCAGUUGGU ((-----.(.((.(((((((......(((((.......)))))...(((((((.......)))).)))))))((((((((((((....))))))))---))))......))))).))).. ( -39.40) >consensus CU_____UAAUAUAGCGCCGGCCUAGCUCAACUGGUAGAGCAACUGACUUGUAAUCAGUAGGUUGGGGGUUCAAGUCCUCUGGCCGGCACCAUUUU___AGAGCCAUUAGCUCAGUUGGU ................(((((((..((((........))))....((((((.((((...........))))))))))....)))))))............((((.....))))....... (-20.86 = -21.46 + 0.60) # Strand winner: reverse (0.99)

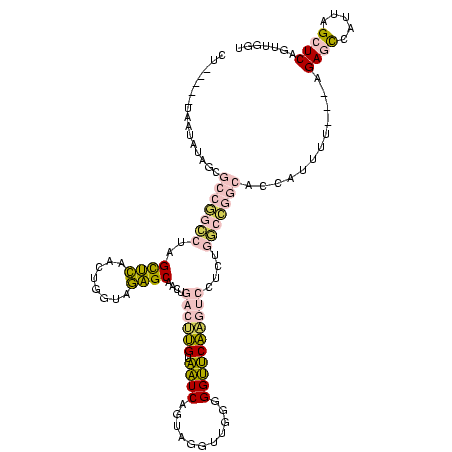
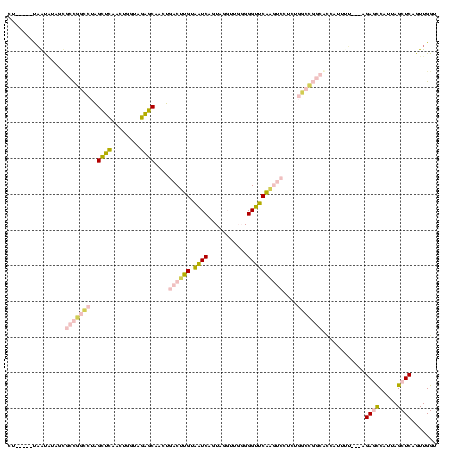
| Location | 1,976,242 – 1,976,358 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.70 |
| Mean single sequence MFE | -36.18 |
| Consensus MFE | -31.28 |
| Energy contribution | -28.56 |
| Covariance contribution | -2.72 |
| Combinations/Pair | 1.27 |
| Mean z-score | -0.70 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1976242 116 + 2809422/720-840 CUCUACCAAUUGAGCUAGGCCGGCAAUAUGUAA----GAAUAAAUGGUGGAGAAUGACGGGUUCGAACCGCCGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAGCUGAGCUAAUUCUCC ((((((((.....(((.....)))..(((....----..)))..))))))))......((((.((......))))))((((((.....)))))).((((........))))......... ( -33.60) >sy_ha.0 1576076 120 - 2685015/720-840 CUCUACCAAUUGAGCUAGGCCGGCAAUAUAUUGUUUUGAUAAAAUGGUGGAGAAUGACGGGUUCGAACCGCCGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAGCUGAGCUAAUUCUCC ((((((((.....(((.....)))....((((.....))))...))))))))......((((.((......))))))((((((.....)))))).((((........))))......... ( -34.00) >sy_sa.0 1551116 113 - 2516575/720-840 CUCUACCAGUUGAGCUAGGCCGGC--UAAAUUA-----AAUAAAUGGUGGAGGAUGACGGGUUCGAACCGCCGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAGCUGAGCUAAUCCUCC ....((((.((((..(((.....)--))..)))-----).....))))(((((((...((((.((......))))))((((((.....)))))).((((........))))..))))))) ( -36.10) >sp_ag.0 2132345 115 - 2160267/720-840 CCGUGACAGGGCGACGUGAUAACCGCUACACUA-----CGGGACCUAUGGGAGUUAACGGGAUCGAACCGCUGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAGCUGAGCUAAACUCCC (((((....((((..((....))))))....))-----))).......(((((((...(((.(((......))))))((((((.....)))))).((((........))))..))))))) ( -38.60) >sp_pn.0 1691504 115 + 2038615/720-840 CCGUGACAGGGCGACGUGAUAACCGCUACACUA-----CGGGACCUUUGGGAGUUAACGGGAUCGAACCGCUGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAGCUGAGCUAAACUCCC (((((....((((..((....))))))....))-----))).......(((((((...(((.(((......))))))((((((.....)))))).((((........))))..))))))) ( -38.60) >consensus CUCUACCAGUUGAGCUAGGCCGGCAAUACAUUA_____AAGAAAUGGUGGAGGAUGACGGGUUCGAACCGCCGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAGCUGAGCUAAUCCUCC (((........)))..................................(((((((...(((.(((......))))))((((((.....)))))).((((........))))..))))))) (-31.28 = -28.56 + -2.72)

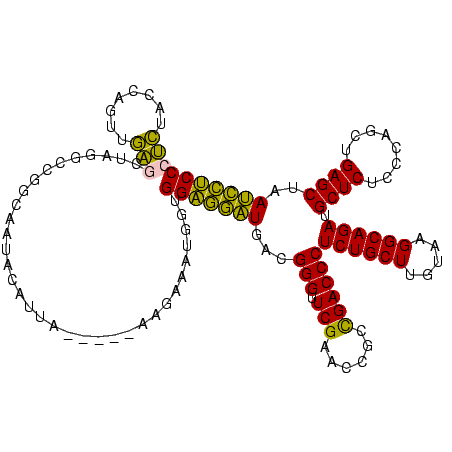
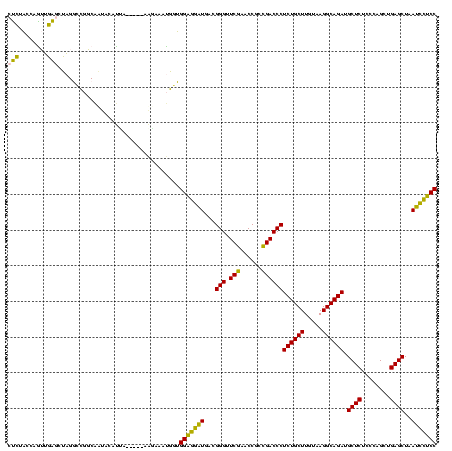
| Location | 1,976,242 – 1,976,358 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.70 |
| Mean single sequence MFE | -36.04 |
| Consensus MFE | -31.38 |
| Energy contribution | -27.78 |
| Covariance contribution | -3.60 |
| Combinations/Pair | 1.30 |
| Mean z-score | -0.14 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1976242 116 + 2809422/720-840 GGAGAAUUAGCUCAGCUGGGAGAGCAUCUGCCUUACAAGCAGAGGGUCGGCGGUUCGAACCCGUCAUUCUCCACCAUUUAUUC----UUACAUAUUGCCGGCCUAGCUCAAUUGGUAGAG ..........(((.((..(..((((.(((((.......)))))(((((((((((..(((.......)))...)))...(((..----....)))..)))))))).))))..)..)).))) ( -35.20) >sy_ha.0 1576076 120 - 2685015/720-840 GGAGAAUUAGCUCAGCUGGGAGAGCAUCUGCCUUACAAGCAGAGGGUCGGCGGUUCGAACCCGUCAUUCUCCACCAUUUUAUCAAAACAAUAUAUUGCCGGCCUAGCUCAAUUGGUAGAG ..........(((.((..(..((((.(((((.......)))))(((((((((((....))).((........))......................)))))))).))))..)..)).))) ( -34.90) >sy_sa.0 1551116 113 - 2516575/720-840 GGAGGAUUAGCUCAGCUGGGAGAGCAUCUGCCUUACAAGCAGAGGGUCGGCGGUUCGAACCCGUCAUCCUCCACCAUUUAUU-----UAAUUUA--GCCGGCCUAGCUCAACUGGUAGAG ..........(((.((..(..((((.(((((.......)))))(((((((((((....))).((........))........-----.......--)))))))).))))..)..)).))) ( -36.90) >sp_ag.0 2132345 115 - 2160267/720-840 GGGAGUUUAGCUCAGCUGGGAGAGCAUCUGCCUUACAAGCAGAGGGUCAGCGGUUCGAUCCCGUUAACUCCCAUAGGUCCCG-----UAGUGUAGCGGUUAUCACGUCGCCCUGUCACGG (((((((.(((...)))((((((((..(((((((........)))).)))..))))..))))...))))))).......(((-----(((.(..(((.......)))..).))...)))) ( -36.30) >sp_pn.0 1691504 115 + 2038615/720-840 GGGAGUUUAGCUCAGCUGGGAGAGCAUCUGCCUUACAAGCAGAGGGUCAGCGGUUCGAUCCCGUUAACUCCCAAAGGUCCCG-----UAGUGUAGCGGUUAUCACGUCGCCCUGUCACGG ((((.((((((...)))((((((((.(((((.......)))))(((((........))))).)))..))))).))).)))).-----..((((((.(((.........)))))).))).. ( -36.90) >consensus GGAGGAUUAGCUCAGCUGGGAGAGCAUCUGCCUUACAAGCAGAGGGUCGGCGGUUCGAACCCGUCAUCCUCCACCAUUUACU_____UAAUAUAGCGCCGGCCUAGCUCAACUGGUAGAG (((((((..((((.(((.....))).(((((.......))))))))).(((((.......))))))))))))..................................(((........))) (-31.38 = -27.78 + -3.60) # Strand winner: reverse (0.73)

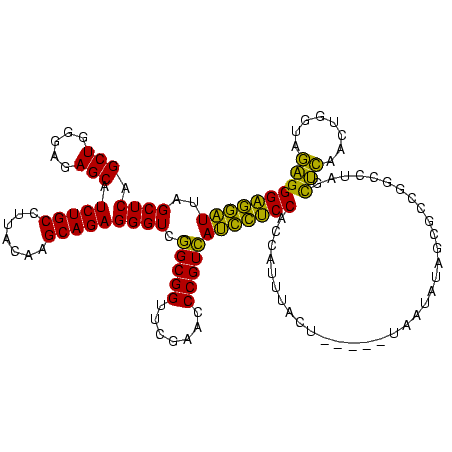
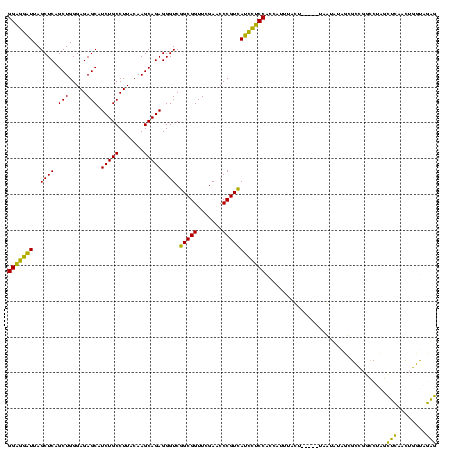
| Location | 1,976,242 – 1,976,362 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.29 |
| Mean single sequence MFE | -35.90 |
| Consensus MFE | -34.88 |
| Energy contribution | -32.64 |
| Covariance contribution | -2.24 |
| Combinations/Pair | 1.31 |
| Mean z-score | -0.60 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1976242 120 + 2809422/760-880 UAAAUGGUGGAGAAUGACGGGUUCGAACCGCCGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAGCUGAGCUAAUUCUCCGAUUUAAAACUGCCUGGCAACGUUCUACUCUAGCGGAACG (((((..((((((((...((((.((......))))))((((((.....)))))).((((........))))..)))))))))))))....(((...))).(((((..(....)..))))) ( -36.30) >sy_ha.0 1576076 119 - 2685015/760-880 AAAAUGGUGGAGAAUGACGGGUUCGAACCGCCGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAGCUGAGCUAAUUCUCCAUUAUAUUA-UGCCUGGCAACGUCCUACUCUAGCGGAACG ...((((((((((((...((((.((......))))))((((((.....)))))).((((........))))..))))))))))))....-(((...))).(((((.........)).))) ( -36.90) >sy_sa.0 1551116 118 - 2516575/760-880 UAAAUGGUGGAGGAUGACGGGUUCGAACCGCCGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAGCUGAGCUAAUCCUCCA-UAUAA-AUUGCCUGGCAACGUCCUACUCUUGCGGAACG ......(((((((((...((((.((......))))))((((((.....)))))).((((........))))..))))))))-)....-.((((...))))(((((.........)).))) ( -37.40) >sp_ag.0 2132345 112 - 2160267/760-880 GGACCUAUGGGAGUUAACGGGAUCGAACCGCUGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAGCUGAGCUAAACUCCCUUU--------GCUAAGCGACUACCUUAUCUCACAGGGGG ...(((..(((((((...(((.(((......))))))((((((.....)))))).((((........))))..))))))).((--------((...))))................))). ( -32.90) >sp_pn.0 1691504 113 + 2038615/760-880 GGACCUUUGGGAGUUAACGGGAUCGAACCGCUGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAGCUGAGCUAAACUCCCUAGA-------GCUAAGCGACUUCCCUAUCUCACAGGGGG ....(((((((((((...(((.(((......))))))((((((.....)))))).((((........))))..))))))).)))-------)..........(((((.......))))). ( -36.00) >consensus GAAAUGGUGGAGGAUGACGGGUUCGAACCGCCGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAGCUGAGCUAAUCCUCCAAUAUA__A_UGCCUGGCAACGUCCUACUCUAGCGGAACG ........(((((((...(((.(((......))))))((((((.....)))))).((((........))))..)))))))....................((((((........)))))) (-34.88 = -32.64 + -2.24)

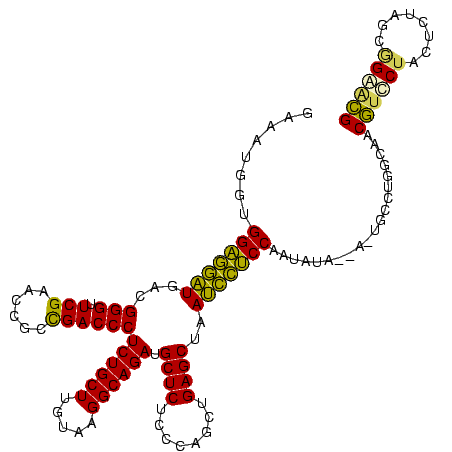
| Location | 1,976,242 – 1,976,358 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.51 |
| Mean single sequence MFE | -27.28 |
| Consensus MFE | -19.82 |
| Energy contribution | -17.30 |
| Covariance contribution | -2.52 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1976242 116 + 2809422/880-1000 UAAGUUC-GACUACCAUCGACGCUAAGGAGCUUAACUUCUGUGUUCGGCAUGGGAACAGGUGUGACCUCCUUGCUAUAUUCACCAGACAUAUGAAUGUA---AUUUAUACAUUCAAAACU ...((.(-((......)))))((.((((((.(((...(((((.(((......))))))))..))).)))))))).................((((((((---.....))))))))..... ( -26.90) >sy_ha.0 1576076 113 - 2685015/880-1000 UAAAUCC-AACUACCAUCGGCGCUAAGGAGCUUAACUUCUGUGUUCGGCAUGGGAACAGGUGUGACCUCCUUGCCAUUGUCACCAGACAAGAGAAUGA------UUAUACAUUCAAAACU .......-........((((((...(((((.(((...(((((.(((......))))))))..))).))))))))).(((((....)))))))(((((.------.....)))))...... ( -27.60) >sy_sa.0 1551116 119 - 2516575/880-1000 UAAGUCC-GACUACCAUCGGCGCUAAGGAGCUUAACUUCUGUGUUCGGCAUGGGAACAGGUGUGACCUCCUUGCCAUUGUCACCAGACAAUGAAAUGUAUGAAAUUAUACAUUCAAAACU ...(.((-((......)))))((.((((((.(((...(((((.(((......))))))))..))).))))))))(((((((....))))))).((((((((....))))))))....... ( -33.60) >sp_ag.0 2132345 113 - 2160267/880-1000 CAACCCCCAACUACUUCCGGCGUUCUAGGGCUUAACUUCUGUGUUCGGCAUGAGAACAGGUGUAUCUCCUAGGCAAUUAUCACUUAACUAUUGAGCCU-------UAUUCACUCAAAAUU ..................((((((((...(((.(((......))).)))...)))))(((((.((..(....)..))...))))).........))).-------............... ( -20.50) >sp_pn.0 1691504 112 + 2038615/880-1000 CAACCCCCAACUACUUCCGGCGUUCUAGGGCUUAACUUCUGUGUUCGGCAUGGGUACAGGUGUAUCUCCUAGGCUAUCGUCACUUAACU-CUGAGUAA-------UACCUACUCAAAAUU ..................((((.(((((((..((...((((((..(.....)..))))))..))..)))))))....))))........-.((((((.-------....))))))..... ( -27.80) >consensus UAACUCC_AACUACCAUCGGCGCUAAGGAGCUUAACUUCUGUGUUCGGCAUGGGAACAGGUGUGACCUCCUUGCCAUUGUCACCAGACAAUUGAAUGU______UUAUACAUUCAAAACU .....................((.((((((.(((...(((((.(((.....))).)))))..))).))))))))..................(((((............)))))...... (-19.82 = -17.30 + -2.52)


Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:54:46 2006