
| Sequence ID | sy_sa.0 |
|---|---|
| Location | 1,780,633 – 1,780,752 |
| Length | 119 |
| Max. P | 0.998953 |

| Location | 1,780,633 – 1,780,752 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.53 |
| Mean single sequence MFE | -26.00 |
| Consensus MFE | -22.39 |
| Energy contribution | -22.17 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 1780633 119 + 2516575/40-160 AAAUAAAUCAUUCGCACUAGGGGUGUUUUGUGACUGAGAUGAGGUAUACCCUCAAACCCUUUGAACCUGAUCUAGAUUAAACUAGCGUAGGAAAGUGUAUUGCGA-UAUCACUGAGUAAC ...........((((((.(((((((((....))).....(((((.....))))).)))))).)..((((..((((......))))..)))).........)))))-.............. ( -27.40) >sy_au.0 1711988 111 - 2809422/40-160 AAAUAAAUCGCACACACUAGGGGUGUUUUAU-ACUGAGAUGAGGCUUGCCCUCAAACCCUUUGAACCUGAUCUAGCUUGAACUAGCGUAGGAAAGUG--UUACUA-UACAUAUG-----U .........((((...(.(((((((((((..-...)))))((((.....))))..)))))).)..((((..((((......))))..))))...)))--).....-........-----. ( -23.90) >sy_ha.0 1925530 114 + 2685015/40-160 AAAUAUA--AAAUGCACUAGGGGUGUGU-AU-ACUGAGAUGAGGUUUAACCUCGAACCCUUUGAACCUGAACUAGUUAAAACUAGCGUAGGAAAGUG--UUGCUAAUAUAAAUAAUUGUG ..((((.--....(((((((((((((..-..-)).....(((((.....))))).))))))....((((..(((((....)))))..))))..))))--).....))))........... ( -26.70) >consensus AAAUAAAUCAAACGCACUAGGGGUGUUUUAU_ACUGAGAUGAGGUUUACCCUCAAACCCUUUGAACCUGAUCUAGAUUAAACUAGCGUAGGAAAGUG__UUGCUA_UAUAAAUGA_U__C ..............((((((((((((......)).....(((((.....))))).))))))....((((..((((......))))..))))..))))....................... (-22.39 = -22.17 + -0.22)

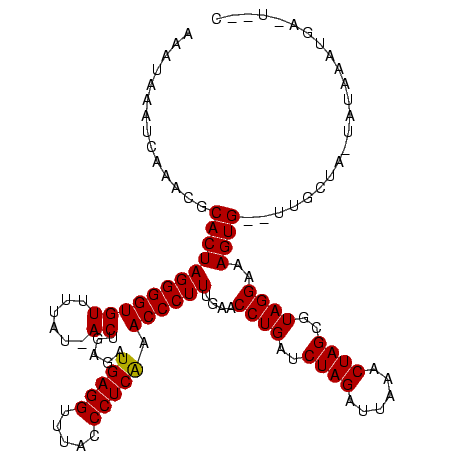
| Location | 1,780,633 – 1,780,752 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.53 |
| Mean single sequence MFE | -23.63 |
| Consensus MFE | -18.50 |
| Energy contribution | -18.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.533027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 1780633 119 + 2516575/40-160 GUUACUCAGUGAUA-UCGCAAUACACUUUCCUACGCUAGUUUAAUCUAGAUCAGGUUCAAAGGGUUUGAGGGUAUACCUCAUCUCAGUCACAAAACACCCCUAGUGCGAAUGAUUUAUUU (((((...))))).-(((((.........(((...((((......))))...))).....((((..(((((.....))))).....((......))..))))..)))))........... ( -24.70) >sy_au.0 1711988 111 - 2809422/40-160 A-----CAUAUGUA-UAGUAA--CACUUUCCUACGCUAGUUCAAGCUAGAUCAGGUUCAAAGGGUUUGAGGGCAAGCCUCAUCUCAGU-AUAAAACACCCCUAGUGUGUGCGAUUUAUUU .-----........-..((((--((((..(((...(((((....)))))...))).....((((..(((((.....))))).....((-.....))..))))))))).)))......... ( -24.30) >sy_ha.0 1925530 114 + 2685015/40-160 CACAAUUAUUUAUAUUAGCAA--CACUUUCCUACGCUAGUUUUAACUAGUUCAGGUUCAAAGGGUUCGAGGUUAAACCUCAUCUCAGU-AU-ACACACCCCUAGUGCAUUU--UAUAUUU .................(((.--......(((..((((((....))))))..))).....((((...((((.....))))......((-..-..))..))))..)))....--....... ( -21.90) >consensus A__A_UCAUUUAUA_UAGCAA__CACUUUCCUACGCUAGUUUAAACUAGAUCAGGUUCAAAGGGUUUGAGGGUAAACCUCAUCUCAGU_AUAAAACACCCCUAGUGCGUAUGAUUUAUUU .......................((((..(((...((((......))))...))).....((((...((((.....))))......((......))..)))))))).............. (-18.50 = -18.50 + 0.00) # Strand winner: forward (1.00)

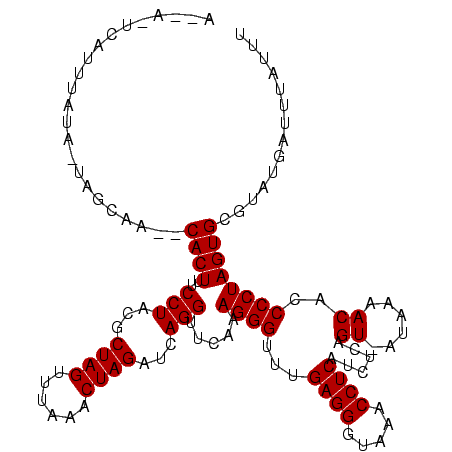

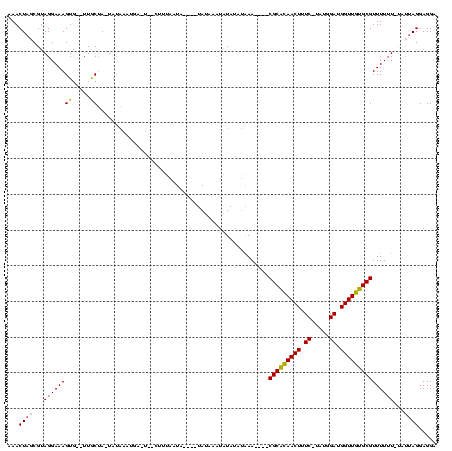
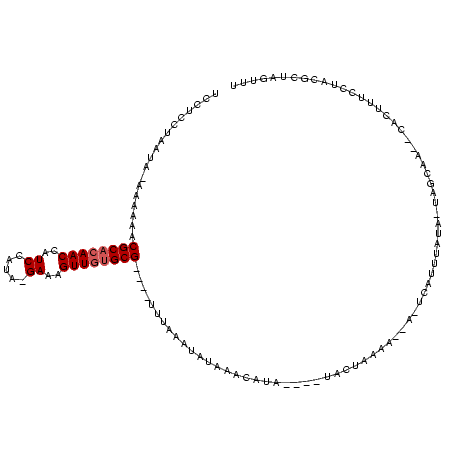
| Location | 1,780,633 – 1,780,745 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.67 |
| Mean single sequence MFE | -28.83 |
| Consensus MFE | -14.63 |
| Energy contribution | -14.63 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.51 |
| SVM decision value | 3.30 |
| SVM RNA-class probability | 0.998953 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 1780633 112 + 2516575/118-238 AAACUAGCGUAGGAAAGUGUAUUGCGA-UAUCACUGAGUAACUUUGGUAUCCGUAUGCCUAUAUAAAAA----CGCGCAACUUUC---UAGAAGGUUGUGCGUUUUUUAUAUUAGGAGGA ..(((((.((.....(((((((....)-)).)))).....)).))))).(((.....((((((((((((----(((((((((((.---...))))))))))).)))))))).)))).))) ( -34.30) >sy_au.0 1711988 101 - 2809422/118-238 GAACUAGCGUAGGAAAGUG--UUACUA-UACAUAUG-----UUUUACUA-----AUAUAUAU-UGUGAA----CGCAUAACUUUCCUAUGGAUGGUUGUGCGUUUUUU-UAUUAGGAGGA ...((((.((((((..(((--(.....-.))))...-----))))))..-----........-...(((----(((((((((.(((...))).))))))))))))...-..))))..... ( -25.40) >sy_ha.0 1925530 112 + 2685015/118-238 AAACUAGCGUAGGAAAGUG--UUGCUAAUAUAAAUAAUUGUGGUAAAUACAUAUAGCAUUUUAUUUAAAUUCACGCACAACUUUC-UACGGAUGGUUGUGCGUUUUUU-----AGGAGGA ....(((((..........--.)))))...(((((((.(((.(((......))).)))..))))))).....((((((((((.((-....)).)))))))))).....-----....... ( -26.80) >consensus AAACUAGCGUAGGAAAGUG__UUGCUA_UAUAAAUGA_U__CUUUAAUA____UAUAAAUAUAUAUAAA____CGCACAACUUUC_UAUGGAUGGUUGUGCGUUUUUU_UAUUAGGAGGA ...((((...((((((((.....))................................................(((((((((.((.....)).)))))))))))))))...))))..... (-14.63 = -14.63 + 0.01)

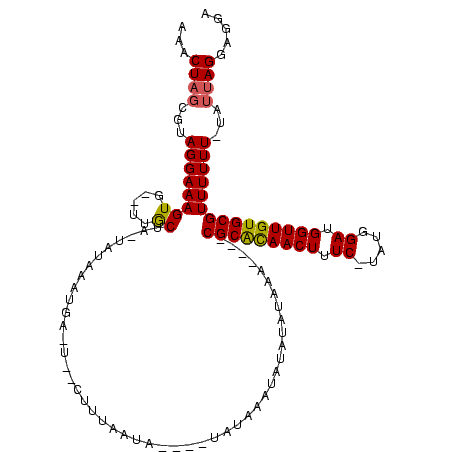
| Location | 1,780,633 – 1,780,745 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.67 |
| Mean single sequence MFE | -18.91 |
| Consensus MFE | -8.90 |
| Energy contribution | -9.57 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878450 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
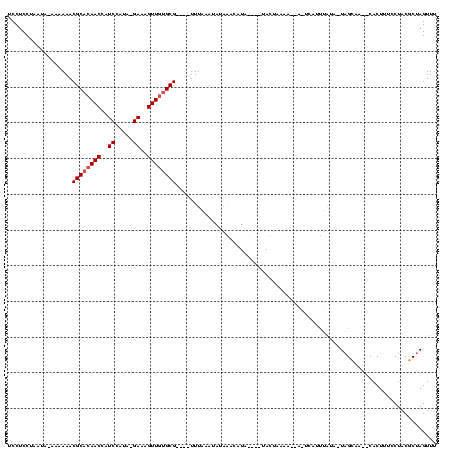
>sy_sa.0 1780633 112 + 2516575/118-238 UCCUCCUAAUAUAAAAAACGCACAACCUUCUA---GAAAGUUGCGCG----UUUUUAUAUAGGCAUACGGAUACCAAAGUUACUCAGUGAUA-UCGCAAUACACUUUCCUACGCUAGUUU ........(((((((((.(((.((((.((...---.)).)))).)))----))))))))).(((.((.(((.....((((......(((...-.))).....))))))))).)))..... ( -21.10) >sy_au.0 1711988 101 - 2809422/118-238 UCCUCCUAAUA-AAAAAACGCACAACCAUCCAUAGGAAAGUUAUGCG----UUCACA-AUAUAUAU-----UAGUAAAA-----CAUAUGUA-UAGUAA--CACUUUCCUACGCUAGUUC ...........-....................(((((((((.....(----((.((.-((((((((-----........-----.)))))))-).))))--))))))))))......... ( -14.70) >sy_ha.0 1925530 112 + 2685015/118-238 UCCUCCU-----AAAAAACGCACAACCAUCCGUA-GAAAGUUGUGCGUGAAUUUAAAUAAAAUGCUAUAUGUAUUUACCACAAUUAUUUAUAUUAGCAA--CACUUUCCUACGCUAGUUU ......(-----(((..(((((((((..((....-))..)))))))))...))))....((((((.....))))))...............((((((..--...........)))))).. ( -20.92) >consensus UCCUCCUAAUA_AAAAAACGCACAACCAUCCAUA_GAAAGUUGUGCG____UUUAAAUAUAAACAUA____UACUAAAA__A_UCAUUUAUA_UAGCAA__CACUUUCCUACGCUAGUUU ..................((((((((..((.....))..))))))))......................................................................... ( -8.90 = -9.57 + 0.67) # Strand winner: forward (1.00)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:54:32 2006