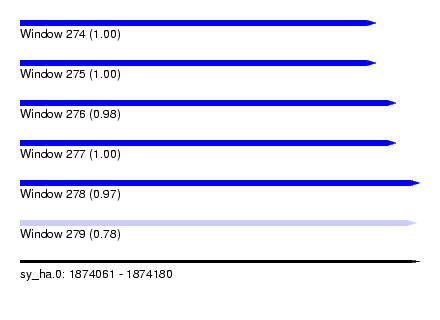
| Sequence ID | sy_ha.0 |
|---|---|
| Location | 1,874,061 – 1,874,180 |
| Length | 119 |
| Max. P | 0.999933 |

| Location | 1,874,061 – 1,874,167 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.79 |
| Mean single sequence MFE | -24.77 |
| Consensus MFE | -13.79 |
| Energy contribution | -13.90 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.87 |
| Structure conservation index | 0.56 |
| SVM decision value | 3.26 |
| SVM RNA-class probability | 0.998865 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1874061 106 + 2685015/0-120 UCACUAUUUAUUGUUAACUUUUUUUCA-------UAAAAAAAGACUUCCAUCCC----AAAACU--GGGACGAAAGUC-UUUCCGUGGUACCACCCAAAUUUAUGCAUAUGCAUACACUU ...........................-------.....((((((((.(.((((----(....)--)))).).)))))-)))..(((....))).......(((((....)))))..... ( -22.20) >sy_au.0 1653908 114 - 2809422/0-120 UUUCUAUUUAUCAUAUUUGUUGUUUAAC------UAAAAAAAGACUUUCAUCCUCUUAAAAAAUAAGGGACGAAAGUCAUUUCCGUGGUACCACCCAAAUUUAUGCCUAAGCAUACACUU .................(((((((((..------........(((((((.(((.((((.....))))))).)))))))......(((....))).............)))))).)))... ( -23.40) >sy_sa.0 1726369 113 + 2516575/0-120 UUUUAAGUUGUUAUAUUUAUAUUUUAAAAUUGCAUAAAAAAAGACUUCCAUCCCUU--AAAACU--GGGACGAA-GUC--UUCCGUGGUACCACCCAAAUUUAUGCCGUAGCAUACACUU ......((((((((.((((.....))))...(((((((..((((((((..((((..--......--)))).)))-)))--))..(((....))).....))))))).)))))).)).... ( -28.70) >consensus UUUCUAUUUAUUAUAUUUAUUUUUUAA_______UAAAAAAAGACUUCCAUCCC_U__AAAACU__GGGACGAAAGUC_UUUCCGUGGUACCACCCAAAUUUAUGCCUAAGCAUACACUU ..........................................(((((.(.((((............)))).).)))))......(((....))).......(((((....)))))..... (-13.79 = -13.90 + 0.11)

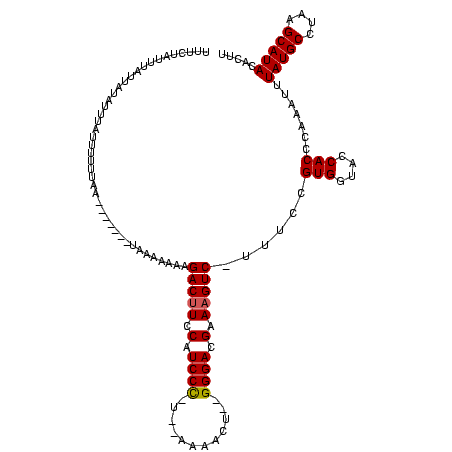
| Location | 1,874,061 – 1,874,167 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.79 |
| Mean single sequence MFE | -31.80 |
| Consensus MFE | -25.15 |
| Energy contribution | -26.77 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.87 |
| Structure conservation index | 0.79 |
| SVM decision value | 4.64 |
| SVM RNA-class probability | 0.999933 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1874061 106 + 2685015/0-120 AAGUGUAUGCAUAUGCAUAAAUUUGGGUGGUACCACGGAAA-GACUUUCGUCCC--AGUUUU----GGGAUGGAAGUCUUUUUUUA-------UGAAAAAAAGUUAACAAUAAAUAGUGA ((((.(((((....))))).))))..(((....)))(((((-((((((((((((--......----)))))))))))))))))...-------........................... ( -33.00) >sy_au.0 1653908 114 - 2809422/0-120 AAGUGUAUGCUUAGGCAUAAAUUUGGGUGGUACCACGGAAAUGACUUUCGUCCCUUAUUUUUUAAGAGGAUGAAAGUCUUUUUUUA------GUUAAACAACAAAUAUGAUAAAUAGAAA .....(((((....))))).(((((.(((....)))(((((.(((((((((((((((.....)))).))))))))))).)))))..------.........))))).............. ( -31.70) >sy_sa.0 1726369 113 + 2516575/0-120 AAGUGUAUGCUACGGCAUAAAUUUGGGUGGUACCACGGAA--GAC-UUCGUCCC--AGUUUU--AAGGGAUGGAAGUCUUUUUUUAUGCAAUUUUAAAAUAUAAAUAUAACAACUUAAAA ((((((.(((....)))((((.(((.(((....)))((((--(((-((((((((--......--..))))).))))))))))......))).)))).............)).)))).... ( -30.70) >consensus AAGUGUAUGCUUAGGCAUAAAUUUGGGUGGUACCACGGAAA_GACUUUCGUCCC__AGUUUU__A_GGGAUGGAAGUCUUUUUUUA_______UUAAAAAAAAAAUAUAAUAAAUAGAAA ((((.(((((....))))).))))..(((....)))(((((.((((((((((((((........)))))))))))))).))))).................................... (-25.15 = -26.77 + 1.61) # Strand winner: reverse (1.00)

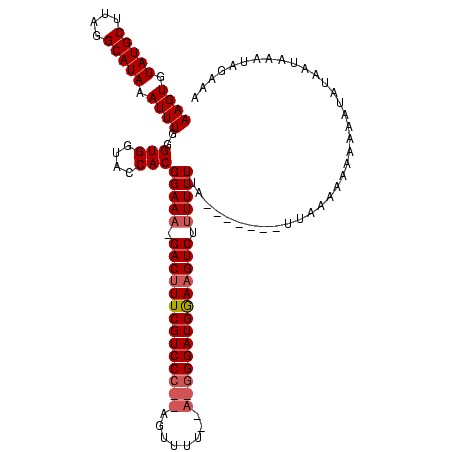
| Location | 1,874,061 – 1,874,173 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.79 |
| Mean single sequence MFE | -28.24 |
| Consensus MFE | -17.31 |
| Energy contribution | -17.87 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.977133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
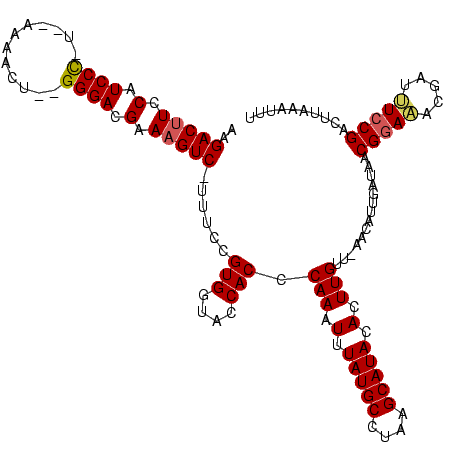
>sy_ha.0 1874061 112 + 2685015/40-160 AAGACUUCCAUCCC----AAAACU--GGGACGAAAGUC-UUUCCGUGGUACCACCCAAAUUUAUGCAUAUGCAUACACUUGUA-AACGUUGCUAACGAAGACGUACUCCGACUUAAAUUU (((((((.(.((((----(....)--)))).).)))))-))..((..((((..........(((((....))))).....((.-..(((.....)))...))))))..)).......... ( -27.50) >sy_au.0 1653908 120 - 2809422/40-160 AAGACUUUCAUCCUCUUAAAAAAUAAGGGACGAAAGUCAUUUCCGUGGUACCACCCAAAUUUAUGCCUAAGCAUACACUUGUUUAAUAUUGAUAACGGAUUGUCUUUCCGACUUAAAUCU ..(((((((.(((.((((.....))))))).)))))))...(((((((((.............))))((((((......)))))).........)))))..(((.....)))........ ( -28.82) >sy_sa.0 1726369 112 + 2516575/40-160 AAGACUUCCAUCCCUU--AAAACU--GGGACGAA-GUC--UUCCGUGGUACCACCCAAAUUUAUGCCGUAGCAUACACUUGUU-GUCAUUGAUAACGGAAAUGAUUUCCGACUUAAAUUU ((((((((..((((..--......--)))).)))-)))--))..(((....))).......(((((....))))).....(((-(((...))))))((((.....))))........... ( -28.40) >consensus AAGACUUCCAUCCC_U__AAAACU__GGGACGAAAGUC_UUUCCGUGGUACCACCCAAAUUUAUGCCUAAGCAUACACUUGUU_AACAUUGAUAACGGAAACGAUUUCCGACUUAAAUUU ..(((((.(.((((............)))).).)))))......(((....))).(((.(.(((((....))))).).)))..............(((((.....))))).......... (-17.31 = -17.87 + 0.56)
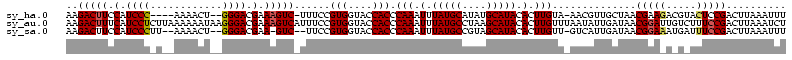
| Location | 1,874,061 – 1,874,173 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.79 |
| Mean single sequence MFE | -38.67 |
| Consensus MFE | -32.23 |
| Energy contribution | -33.40 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.09 |
| Mean z-score | -4.19 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.96 |
| SVM RNA-class probability | 0.997907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1874061 112 + 2685015/40-160 AAAUUUAAGUCGGAGUACGUCUUCGUUAGCAACGUU-UACAAGUGUAUGCAUAUGCAUAAAUUUGGGUGGUACCACGGAAA-GACUUUCGUCCC--AGUUUU----GGGAUGGAAGUCUU .........(((..((((.....(((.....)))..-..(((((.(((((....))))).)))))....))))..))).((-((((((((((((--......----)))))))))))))) ( -38.50) >sy_au.0 1653908 120 - 2809422/40-160 AGAUUUAAGUCGGAAAGACAAUCCGUUAUCAAUAUUAAACAAGUGUAUGCUUAGGCAUAAAUUUGGGUGGUACCACGGAAAUGACUUUCGUCCCUUAUUUUUUAAGAGGAUGAAAGUCUU ........(((.....)))..(((((......(((((..(((((.(((((....))))).)))))..)))))..)))))...(((((((((((((((.....)))).))))))))))).. ( -40.90) >sy_sa.0 1726369 112 + 2516575/40-160 AAAUUUAAGUCGGAAAUCAUUUCCGUUAUCAAUGAC-AACAAGUGUAUGCUACGGCAUAAAUUUGGGUGGUACCACGGAA--GAC-UUCGUCCC--AGUUUU--AAGGGAUGGAAGUCUU ........(((((((.....))))(....)...)))-..(((((.(((((....))))).))))).(((....)))..((--(((-((((((((--......--..))))).)))))))) ( -36.60) >consensus AAAUUUAAGUCGGAAAACAUAUCCGUUAUCAAUGUU_AACAAGUGUAUGCUUAGGCAUAAAUUUGGGUGGUACCACGGAAA_GACUUUCGUCCC__AGUUUU__A_GGGAUGGAAGUCUU ..........(((((.....)))))..............(((((.(((((....))))).))))).(((....)))......((((((((((((((........)))))))))))))).. (-32.23 = -33.40 + 1.17) # Strand winner: reverse (1.00)

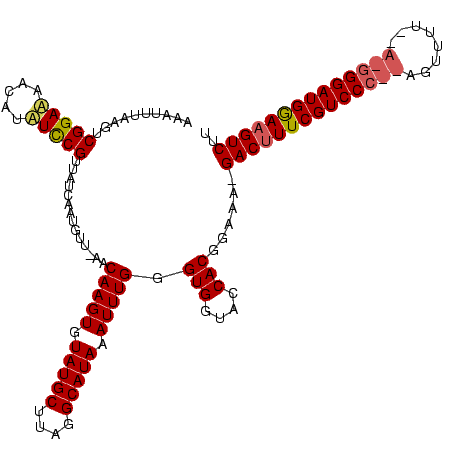
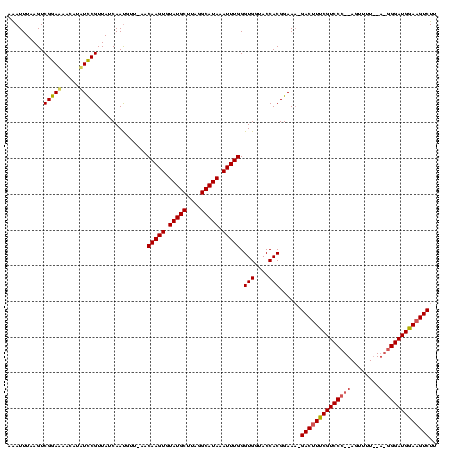
| Location | 1,874,061 – 1,874,180 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.78 |
| Mean single sequence MFE | -29.17 |
| Consensus MFE | -25.15 |
| Energy contribution | -24.93 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.967832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1874061 119 + 2685015/80-200 AACAUGUAUUAGUUUCUUUCAUCUUUUUGGUUACUUAAAGAAAUUUAAGUCGGAGUACGUCUUCGUUAGCAACGUU-UACAAGUGUAUGCAUAUGCAUAAAUUUGGGUGGUACCACGGAA ...........(((.((..(............(((((((....))))))).((((.....)))))..)).)))...-..(((((.(((((....))))).))))).(((....))).... ( -24.00) >sy_au.0 1653908 119 - 2809422/80-200 AAU-UAUUUAAAGUCUUUUCACCUUUUUGGUUACUUAAAGAGAUUUAAGUCGGAAAGACAAUCCGUUAUCAAUAUUAAACAAGUGUAUGCUUAGGCAUAAAUUUGGGUGGUACCACGGAA ...-........(((((((((((.....))).(((((((....))))))).))))))))..(((((......(((((..(((((.(((((....))))).)))))..)))))..))))). ( -33.90) >sy_sa.0 1726369 118 + 2516575/80-200 GAUGUA-GUUAGUUUUUUUCACCUUUUUGGUUACUUAAAGAAAUUUAAGUCGGAAAUCAUUUCCGUUAUCAAUGAC-AACAAGUGUAUGCUACGGCAUAAAUUUGGGUGGUACCACGGAA ......-........((((((((.....))).(((((((....))))))).)))))....(((((((((((.....-..(((((.(((((....))))).)))))..)))))..)))))) ( -29.60) >consensus AAU_UAUAUUAGUUUUUUUCACCUUUUUGGUUACUUAAAGAAAUUUAAGUCGGAAAACAUAUCCGUUAUCAAUGUU_AACAAGUGUAUGCUUAGGCAUAAAUUUGGGUGGUACCACGGAA ..............(((((((((.....))).(((((((....))))))).))))))....(((((.............(((((.(((((....))))).)))))((.....))))))). (-25.15 = -24.93 + -0.21) # Strand winner: reverse (1.00)

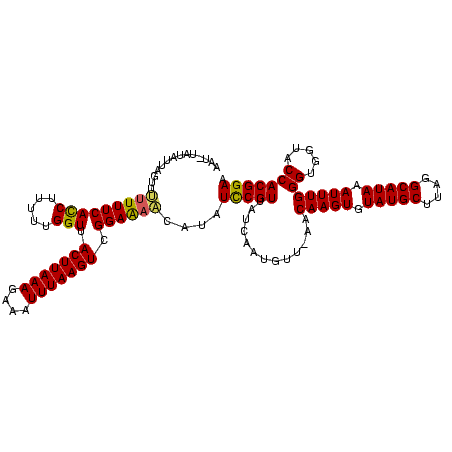
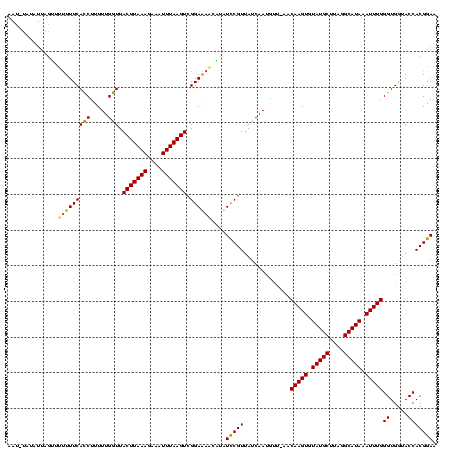
| Location | 1,874,061 – 1,874,179 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.42 |
| Mean single sequence MFE | -21.96 |
| Consensus MFE | -14.74 |
| Energy contribution | -13.53 |
| Covariance contribution | -1.21 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1874061 118 + 2685015/120-240 GUA-AACGUUGCUAACGAAGACGUACUCCGACUUAAAUUUCUUUAAGUAACCAAAAAGAUGAAAGAAACUAAUACAUGUUGGAGUAAAUUGCAGUCACGUUUACUCUCCCUGUA-CAACA (((-(((((.(((..(((.....(((((((((.....(((((((.................))))))).........)))))))))..))).))).))))))))..........-..... ( -24.77) >sy_au.0 1653908 119 - 2809422/120-240 GUUUAAUAUUGAUAACGGAUUGUCUUUCCGACUUAAAUCUCUUUAAGUAACCAAAAAGGUGAAAAGACUUUAAAUA-AUUGGAAUAACUUGCAGUCACGAUUAUUCUCCCUAAAACAAUU ((((......(((((((((((((..((((((((((((....))))))).(((.....)))................-..)))))......)))))).)).))))).......)))).... ( -19.32) >sy_sa.0 1726369 118 + 2516575/120-240 GUU-GUCAUUGAUAACGGAAAUGAUUUCCGACUUAAAUUUCUUUAAGUAACCAAAAAGGUGAAAAAAACUAAC-UACAUCGGAUUAUCUUUCAGUCGUGGAUAAUCUCCCUUUUACUUCU (((-(((...))))))((((.....)))).(((((((....))))))).....((((((.((..........(-......)((((((((.........))))))))))))))))...... ( -21.80) >consensus GUU_AACAUUGAUAACGGAAACGAUUUCCGACUUAAAUUUCUUUAAGUAACCAAAAAGGUGAAAAAAACUAAAAUA_AUUGGAAUAACUUGCAGUCACGAUUAAUCUCCCUAUAACAACU ......((((.....(((((.....)))))(((((((....))))))).........))))...................(((((((((.........)))))))))............. (-14.74 = -13.53 + -1.21)

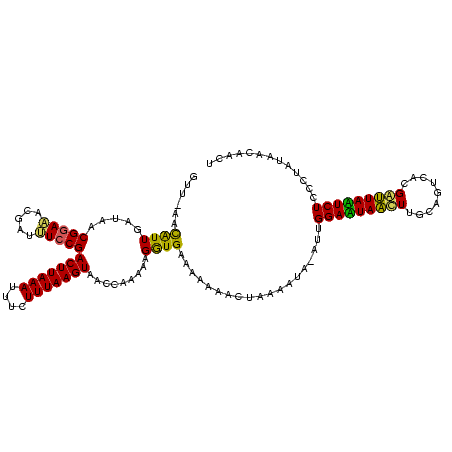
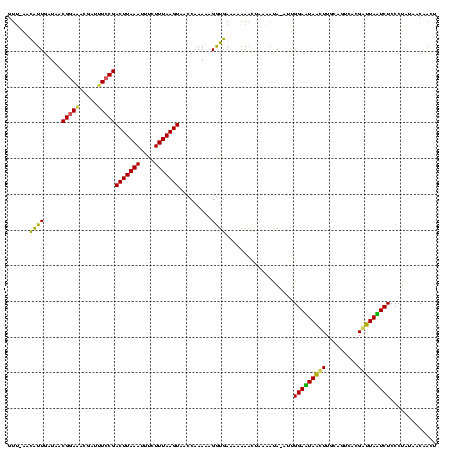
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:54:27 2006