| Sequence ID | sy_ha.0 |
|---|---|
| Location | 878,122 – 878,241 |
| Length | 119 |
| Max. P | 1.000000 |

| Location | 878,122 – 878,241 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.94 |
| Mean single sequence MFE | -29.37 |
| Consensus MFE | -21.38 |
| Energy contribution | -20.83 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.73 |
| SVM decision value | 4.26 |
| SVM RNA-class probability | 0.999852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 878122 119 - 2685015/0-120 AUGGUAUCUAAUUACAAGACAAAAUAAAAAGAACGCCGUCAUAACAGCGUUCUCA-ACCUAAAAAUAUAUGUAAUUUUUAGUUAAUAGCGUCCUGGGAGGGAUUCGAACCCCCGACCGAU .((((.........((.(((.........(((((((.((....)).)))))))..-..((((((((.......))))))))........))).))((.(((.......))))).)))).. ( -27.90) >sy_au.0 1191692 119 + 2809422/0-120 AUACUAUAUUACUGAAAUUCAAAACAAAAAGAGCCCCGUAAUCACGGAACUCUUUUGUUUGGUAAUGCGUAUAAAAAU-ACCUAUAAACGUCCUGGGAGGGAUUCGAACCCCCGACCGAU ((((..((((((..((..........(((((((..((((....))))..))))))).))..)))))).))))......-..........(((..(((.((........)))))))).... ( -27.70) >sy_sa.0 1707858 118 + 2516575/0-120 AUUGUAUCGCAUUGUAG-AUAAAAUAAAAGAAACCCCGUCGUUACGGGGUUUCCUAGUUUGA-AUUGUGCGCUAUUUAACGCUAUUAGUGUCCUGGGAGGGAUUCGAACCCCCGACCGAU ...(((.(((((.....-...((((....((((((((((....))))))))))...))))..-...))))).)))...((((.....))))..(((..(((.........)))..))).. ( -32.52) >consensus AUAGUAUCUAAUUGAAA_ACAAAAUAAAAAGAACCCCGUCAUAACGGAGUUCUCU_GUUUGA_AAUGUGUGUAAUUUU_AGCUAUUAGCGUCCUGGGAGGGAUUCGAACCCCCGACCGAU .............................((((((((((....))))))))))....................................(((..(((.((........)))))))).... (-21.38 = -20.83 + -0.55)

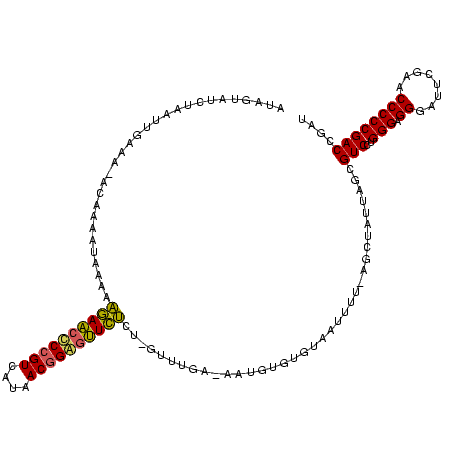
| Location | 878,122 – 878,241 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.94 |
| Mean single sequence MFE | -35.14 |
| Consensus MFE | -27.50 |
| Energy contribution | -24.63 |
| Covariance contribution | -2.87 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.78 |
| SVM decision value | 7.21 |
| SVM RNA-class probability | 1.000000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 878122 119 - 2685015/0-120 AUCGGUCGGGGGUUCGAAUCCCUCCCAGGACGCUAUUAACUAAAAAUUACAUAUAUUUUUAGGU-UGAGAACGCUGUUAUGACGGCGUUCUUUUUAUUUUGUCUUGUAAUUAGAUACCAU ...(((.(((((.......))))).((((((........((((((((.......))))))))..-.(((((((((((....)))))))))))........)))))).........))).. ( -36.90) >sy_au.0 1191692 119 + 2809422/0-120 AUCGGUCGGGGGUUCGAAUCCCUCCCAGGACGUUUAUAGGU-AUUUUUAUACGCAUUACCAAACAAAAGAGUUCCGUGAUUACGGGGCUCUUUUUGUUUUGAAUUUCAGUAAUAUAGUAU ...((..((((((....)))))).))..((.(((((..(((-(.............))))(((((((((((((((((....))))))))).))))))))))))).))............. ( -33.12) >sy_sa.0 1707858 118 + 2516575/0-120 AUCGGUCGGGGGUUCGAAUCCCUCCCAGGACACUAAUAGCGUUAAAUAGCGCACAAU-UCAAACUAGGAAACCCCGUAACGACGGGGUUUCUUUUAUUUUAU-CUACAAUGCGAUACAAU ((((((((((((........)))))...))).......(((((....))))).....-.......((((((((((((....)))))))))))).........-........))))..... ( -35.40) >consensus AUCGGUCGGGGGUUCGAAUCCCUCCCAGGACGCUAAUAGCU_AAAAUUACACACAUU_UCAAAC_AGAGAACCCCGUAAUGACGGGGUUCUUUUUAUUUUGU_UUACAAUAAGAUACAAU ...((..((((((....)))))).))..................................((((..(((((((((((....)))))))))))...))))..................... (-27.50 = -24.63 + -2.87) # Strand winner: reverse (1.00)

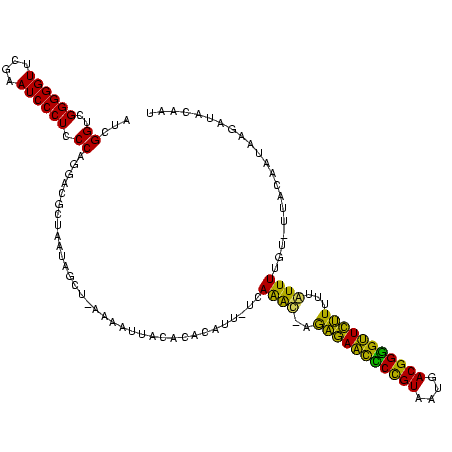
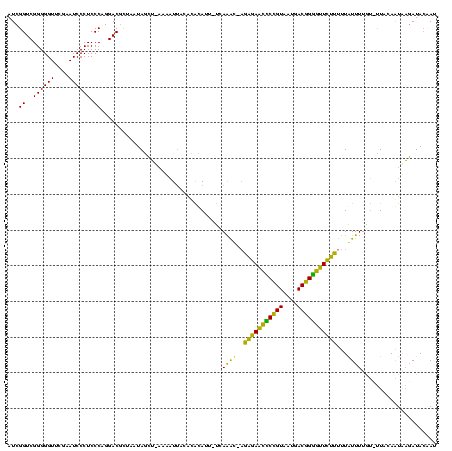
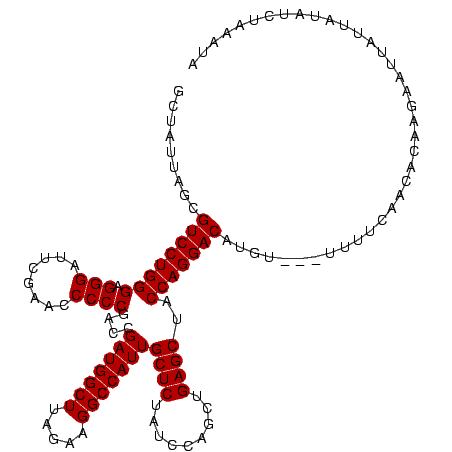
| Location | 878,122 – 878,240 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.43 |
| Mean single sequence MFE | -36.13 |
| Consensus MFE | -31.30 |
| Energy contribution | -31.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 878122 118 - 2685015/80-200 GUUAAUAGCGUCCUGGGAGGGAUUCGAACCCCCGACCGAUGGCUUAGAAGGCCAUUGCUCUAUCCAGCUGAGCUACCAGGACAUGAA--UUUUUAACACAAGAAUUAUUAUAGCUAAAGA .....((((((((((((.(((........))))....(((((((.....)))))))((((.........))))..)))))))...((--(((((.....)))))))......)))).... ( -37.10) >sy_au.0 1191692 117 + 2809422/80-200 CCUAUAAACGUCCUGGGAGGGAUUCGAACCCCCGACCGAUGGCUUAGAAGGCCAUUGCUCUAUCCAGCUGAGCUACCAGGACACGU---UUAACAACACAAGAAUUAUUAUAUCUAAAUG ....(((((((((((((.(((........))))....(((((((.....)))))))((((.........))))..)))))))..))---)))............................ ( -32.50) >sy_sa.0 1707858 120 + 2516575/80-200 GCUAUUAGUGUCCUGGGAGGGAUUCGAACCCCCGACCGAUGGCUUAGAAGGCCAUUGCUCUAUCCAGCUGAGCUACCAGGACAUAUUGUUUUUGAACACAAAAAUAAUUAUAUCUAACUU .......((((((((((.(((........))))....(((((((.....)))))))((((.........))))..)))))))))((((((((((....))))))))))............ ( -38.80) >consensus GCUAUUAGCGUCCUGGGAGGGAUUCGAACCCCCGACCGAUGGCUUAGAAGGCCAUUGCUCUAUCCAGCUGAGCUACCAGGACAUGU___UUUUCAACACAAGAAUUAUUAUAUCUAAAUA .........((((((((.(((........))))....(((((((.....)))))))((((.........))))..)))))))...................................... (-31.30 = -31.30 + 0.00)

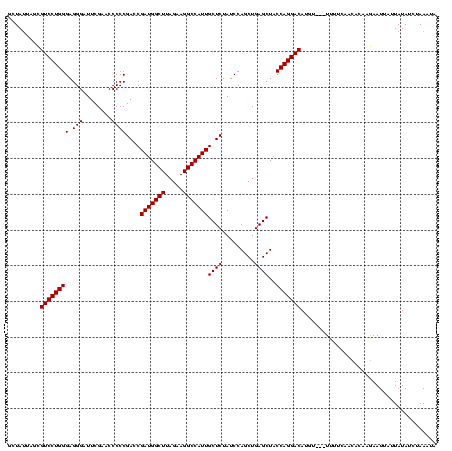
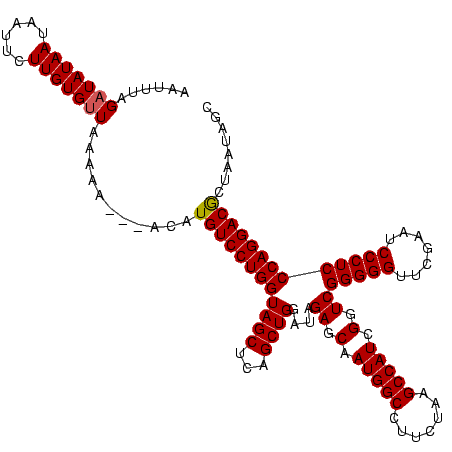
| Location | 878,122 – 878,240 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.43 |
| Mean single sequence MFE | -34.93 |
| Consensus MFE | -32.09 |
| Energy contribution | -32.20 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 878122 118 - 2685015/80-200 UCUUUAGCUAUAAUAAUUCUUGUGUUAAAAA--UUCAUGUCCUGGUAGCUCAGCUGGAUAGAGCAAUGGCCUUCUAAGCCAUCGGUCGGGGGUUCGAAUCCCUCCCAGGACGCUAUUAAC ....((((.....((((......))))....--.....(((((((((((...))))....((.(.(((((.......))))).).))(((((.......))))))))))))))))..... ( -34.00) >sy_au.0 1191692 117 + 2809422/80-200 CAUUUAGAUAUAAUAAUUCUUGUGUUGUUAA---ACGUGUCCUGGUAGCUCAGCUGGAUAGAGCAAUGGCCUUCUAAGCCAUCGGUCGGGGGUUCGAAUCCCUCCCAGGACGUUUAUAGG ....(((((.(((((((......))))))).---....(((((((((((...))))....((.(.(((((.......))))).).))(((((.......))))))))))))))))).... ( -34.10) >sy_sa.0 1707858 120 + 2516575/80-200 AAGUUAGAUAUAAUUAUUUUUGUGUUCAAAAACAAUAUGUCCUGGUAGCUCAGCUGGAUAGAGCAAUGGCCUUCUAAGCCAUCGGUCGGGGGUUCGAAUCCCUCCCAGGACACUAAUAGC ..(((((.....(((.((((((....)))))).))).((((((((((((...))))....((.(.(((((.......))))).).))(((((.......))))))))))))))))))... ( -36.70) >consensus AAUUUAGAUAUAAUAAUUCUUGUGUUAAAAA___ACAUGUCCUGGUAGCUCAGCUGGAUAGAGCAAUGGCCUUCUAAGCCAUCGGUCGGGGGUUCGAAUCCCUCCCAGGACGCUAAUAGC ......(((((((......)))))))...........((((((((((((...))))....((.(.(((((.......))))).).))(((((.......)))))))))))))........ (-32.09 = -32.20 + 0.11) # Strand winner: reverse (0.71)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:54:22 2006