
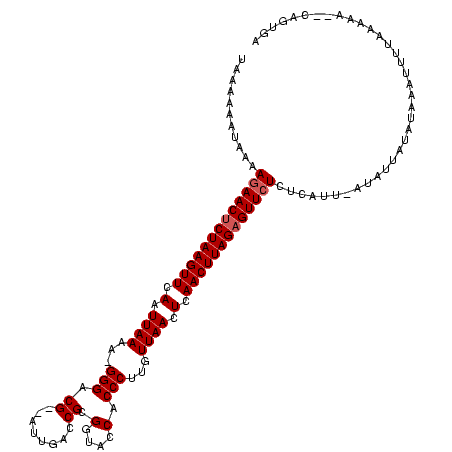
| Sequence ID | sy_ha.0 |
|---|---|
| Location | 1,786,422 – 1,786,539 |
| Length | 117 |
| Max. P | 0.997014 |

| Location | 1,786,422 – 1,786,539 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.99 |
| Mean single sequence MFE | -25.87 |
| Consensus MFE | -16.63 |
| Energy contribution | -17.30 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.64 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.997014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1786422 117 + 2685015/0-120 UUAAAUGUAAAAGAACUCUAAGUUCAAUUAAAAAGGGACGG-AAUUACCGCGGUACCACCCUUGUUAACUCAACUUAGAGUUCUCUCAU--AUAUAAUAUUACUUUGUAAAAAUCAGUGA ....(((....(((((((((((((.(.((((.(((((.(((-.....))).(....).))))).)))).).)))))))))))))..)))--.(((((.......)))))........... ( -28.70) >sy_au.0 1598539 116 - 2809422/0-120 UAAAAAUUUAAAAAACUCUAAGUUAAAUUAAAA-GGGACG--AUUGCUCGCGGUACCACCCUUAUUAACUCAACUUAGAGUUCUCUCAUU-ACAUUAUUUAAAUAUUAAAAAGCCAGUGA .............(((((((((((.(.((((((-(((...--((((....))))....))))).)))).).)))))))))))........-............................. ( -21.20) >sy_sa.0 1650268 117 + 2516575/0-120 UAUACAAUAAAAGAACCCUAAGUUCAAUUAACA-GGGACGAUAUUGACCGCGUUACCACCCUGGUUAACUCAACUUAGAGUUCUCUCAUUUAUGUUAUAUAAAUUGUGACAA--CAGUGA ...........(((((.(((((((.(.((((((-((((((..........))).....)))).))))).).))))))).))))).(((((..(((((((.....))))))).--.))))) ( -27.70) >consensus UAAAAAAUAAAAGAACUCUAAGUUCAAUUAAAA_GGGACG__AUUGACCGCGGUACCACCCUUGUUAACUCAACUUAGAGUUCUCUCAUU_AUAUUAUAUAAAUUUUAAAAA__CAGUGA ...........(((((((((((((.(.((((...(((.((........)).(....).)))...)))).).))))))))))))).................................... (-16.63 = -17.30 + 0.67)

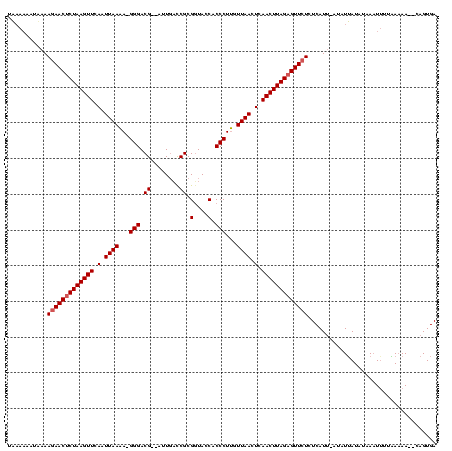
| Location | 1,786,422 – 1,786,539 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.99 |
| Mean single sequence MFE | -38.93 |
| Consensus MFE | -29.50 |
| Energy contribution | -29.18 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -5.57 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.994017 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1786422 117 + 2685015/0-120 UCACUGAUUUUUACAAAGUAAUAUUAUAU--AUGAGAGAACUCUAAGUUGAGUUAACAAGGGUGGUACCGCGGUAAUU-CCGUCCCUUUUUAAUUGAACUUAGAGUUCUUUUACAUUUAA .............................--.(((((((((((((((((.((((((.(((((((((((....)))...-))).))))).)))))).)))))))))))))))))....... ( -40.00) >sy_au.0 1598539 116 - 2809422/0-120 UCACUGGCUUUUUAAUAUUUAAAUAAUGU-AAUGAGAGAACUCUAAGUUGAGUUAAUAAGGGUGGUACCGCGAGCAAU--CGUCCC-UUUUAAUUUAACUUAGAGUUUUUUAAAUUUUUA ..............(((((.....)))))-....((((((((((((((((((((((.(((((((((..(....)..))--)).)))-))))))))))))))))))))))))......... ( -35.30) >sy_sa.0 1650268 117 + 2516575/0-120 UCACUG--UUGUCACAAUUUAUAUAACAUAAAUGAGAGAACUCUAAGUUGAGUUAACCAGGGUGGUAACGCGGUCAAUAUCGUCCC-UGUUAAUUGAACUUAGGGUUCUUUUAUUGUAUA ....((--((((..........))))))..(((((((((((((((((((.(((((((.(((((((((..........))))).)))-)))))))).)))))))))))))))))))..... ( -41.50) >consensus UCACUG__UUUUAAAAAUUUAUAUAAUAU_AAUGAGAGAACUCUAAGUUGAGUUAACAAGGGUGGUACCGCGGUCAAU__CGUCCC_UUUUAAUUGAACUUAGAGUUCUUUUAAUUUUUA ................................(((((((((((((((((.((((((.(((((.......((((......))))))).)))))))).)))))))))))))))))....... (-29.50 = -29.18 + -0.33) # Strand winner: reverse (1.00)


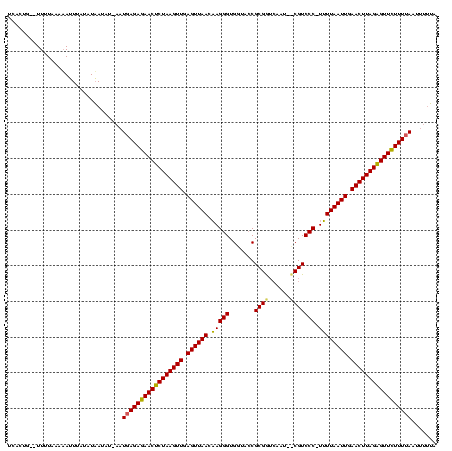
| Location | 1,786,422 – 1,786,539 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.83 |
| Mean single sequence MFE | -22.07 |
| Consensus MFE | -16.51 |
| Energy contribution | -13.30 |
| Covariance contribution | -3.21 |
| Combinations/Pair | 1.44 |
| Mean z-score | -0.84 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1786422 117 + 2685015/78-198 UCAGACAAGUAGGAGAUAAUAAACUUUUUACAGCGAAUUAGG-GUUGGUGAAAGCCUAAUAAAAGUCUAUCUCUUAUAUCACUGAUUUUUACAAAGUAAUAUUAUAU--AUGAGAGAACU ..((((..(((((((.........))))))).....((((((-.((.....)).))))))....)))).((((((((((...((((..((((...)))).)))).))--))))))))... ( -24.60) >sy_au.0 1598539 119 - 2809422/78-198 ACAGACGCGUAAUAUACAUUUAACUUUUCACAGCGAAUUAGGUAAUGGUGAGAGCCUAGUAAAAGCAUGUAUGUUAUAUCACUGGCUUUUUAAUAUUUAAAUAAUGU-AAUGAGAGAACU .(((....(((((((((((....(((((.((.((.......))...(((....)))..))))))).)))))))))))....))).(((((..(((((.....)))))-...))))).... ( -21.20) >sy_sa.0 1650268 117 + 2516575/78-198 ACAGACACGUUUGAAACGAAACACAUAUCAAAGCGAGUUAGG-AAUAGUGAGAGCCUUUCAUAUGUGCGUUUCUAAUAUCACUG--UUGUCACAAUUUAUAUAACAUAAAUGAGAGAACU ........(((.((((((...(((((((.....(......)(-((..((....))..)))))))))))))))).)))......(--((.((...((((((.....))))))..)).))). ( -20.40) >consensus ACAGACACGUAAGAAACAAUAAACUUUUCACAGCGAAUUAGG_AAUGGUGAGAGCCUAAUAAAAGUAUGUAUCUUAUAUCACUG__UUUUAAAAAUUUAUAUAAUAU_AAUGAGAGAACU .(((....((((((((((....((((((....((..((((.....))))....)).....)))))).))))))))))....))).................................... (-16.51 = -13.30 + -3.21) # Strand winner: reverse (0.83)


Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:54:09 2006