
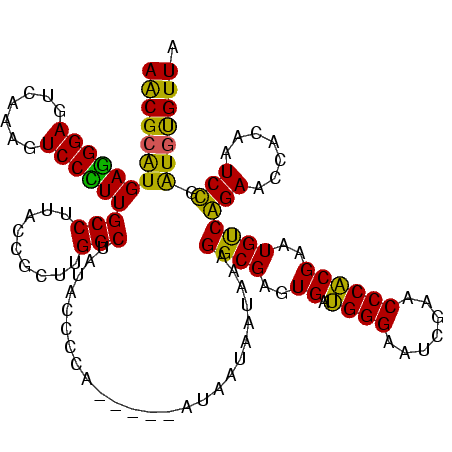
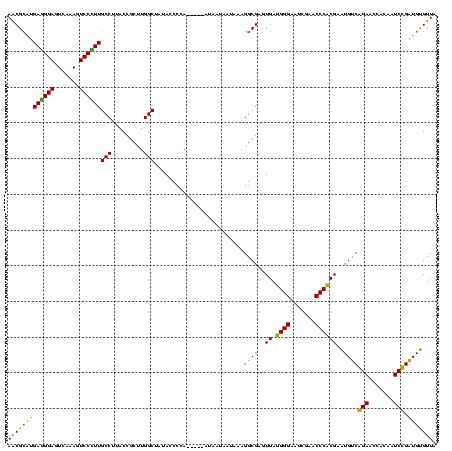
| Sequence ID | sy_ha.0 |
|---|---|
| Location | 1,574,809 – 1,574,929 |
| Length | 120 |
| Max. P | 0.998829 |

| Location | 1,574,809 – 1,574,927 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.76 |
| Mean single sequence MFE | -36.34 |
| Consensus MFE | -29.81 |
| Energy contribution | -27.28 |
| Covariance contribution | -2.52 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1574809 118 - 2685015/0-120 AGCGCGUGACGGAGUCAAAGUCCGUUGCCUUACCGCUUGGCUAUAGCCCAUUA--AUAAAAUUAAGGGCGGUUGAAGGGAAUCGAACCCUCGAGUGUCGGAACCACAAUCCGAUGCGUUA ((((((..(((((.......)))))..).....((((((......((((.(((--((...)))))))))......((((.......))))))))))(((((.......))))).))))). ( -39.00) >sy_sa.0 1549924 115 - 2516575/0-120 AGCGCGUGAUGGAGUCAAAGUCCAUUGCCUUACCGCUUGGCUAUAGCCCA-----AUAAUACUAAGGGCGGAUGAUGGGAAUCGAACCCACGAGUGUCGGAACCACAAUCCGAUGCGUUA ((((((..(((((.......)))))..).....((((((......((((.-----..........))))......((((.......))))))))))(((((.......))))).))))). ( -37.00) >sy_au.0 1974979 115 + 2809422/0-120 AACGAGUGACGGAGUCAAAGUCCGUUGCCUUACCGCUUGGCUAUAGCCCA-----UUAAUAAUAAGGGCGGCUGAAGGGGAUCGAACCCUCGAAUGUCGGAACCACAAUCCGAUGUGUUA .....(..(((((.......)))))..)....((.(((((((...((((.-----(((....))))))))))).))).)).((((....))))((((((((.......)))))))).... ( -36.00) >sp_mu.0 1839095 120 - 2030921/0-120 AACGCAUGAGGGAGUCAAAGUCCCUUGCCUUACCGCUUGGCUAUACCCCAAUAAUAUAAUAAUAUAGGCGAGUGAUGGGGAUCGAACCCACGCAUGCCAGAGCCACAAUCUGGUGUGUUA ...(((..(((((.......)))))))).....((((((.(((((.................))))).)))))).((((.......)))).((((((((((.......)))))))))).. ( -37.93) >sp_pn.0 1850856 114 + 2038615/0-120 AACGCAUGAGGGAGUCAAAGUCCCUUGCCUUACCGCUUGGCUAUACCCCAAUA------UAAAAUAGGCGAGUGAUGGGGAUCGAACCCACGCAUGCCAGAGCCACAAUCUGGUGUGUUA ...(((..(((((.......)))))))).....((((((.((((.........------....)))).)))))).((((.......)))).((((((((((.......)))))))))).. ( -37.32) >sp_ag.0 1904059 110 - 2160267/0-120 AACGCAUGAGGGAGUCAAAGUCCCUUGCCUUACCGCUUGGCUAUACCCCA---------UGAAA-AGGCGAGUGAUGGGAAUCGAACCCACGAAUGUCAGAGCCACAAUCUGAUGUGUUA (((((....(((..((...(((.((((((((....(.(((.......)))---------.)..)-))))))).)))..))......)))......((((((.......))))))))))). ( -30.80) >consensus AACGCAUGAGGGAGUCAAAGUCCCUUGCCUUACCGCUUGGCUAUACCCCA_____AUAAUAAUAAAGGCGAGUGAUGGGAAUCGAACCCACGAAUGUCAGAACCACAAUCCGAUGUGUUA (((((((((((((.......))))))(((.........))).........................((((..((.((((.......))))))..))))(((.......))).))))))). (-29.81 = -27.28 + -2.52)

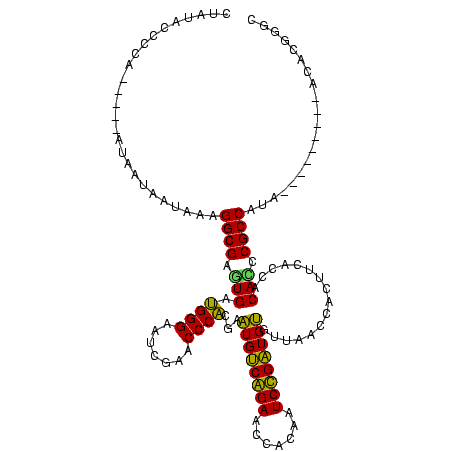
| Location | 1,574,809 – 1,574,918 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.43 |
| Mean single sequence MFE | -33.97 |
| Consensus MFE | -26.83 |
| Energy contribution | -24.78 |
| Covariance contribution | -2.05 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1574809 109 - 2685015/40-160 CUAUAGCCCAUUA--AUAAAAUUAAGGGCGGUUGAAGGGAAUCGAACCCUCGAGUGUCGGAACCACAAUCCGAUGCGUUAACCACUUCGCCACAACCGCCAUG---------GCAGGGAC .....((((.(((--((...)))))))))(((.((((((..((((....))))((((((((.......)))))))).....)).)))))))....((((....---------)).))... ( -36.30) >sy_sa.0 1549924 106 - 2516575/40-160 CUAUAGCCCA-----AUAAUACUAAGGGCGGAUGAUGGGAAUCGAACCCACGAGUGUCGGAACCACAAUCCGAUGCGUUAACCACUUCGCCACAACCGCCAUG---------GCAGGGGC .....((((.-----......(((..(((((.((.(((((((((......)))((((((((.......)))))))).........))).))))).))))).))---------)...)))) ( -34.60) >sy_au.0 1974979 106 + 2809422/40-160 CUAUAGCCCA-----UUAAUAAUAAGGGCGGCUGAAGGGGAUCGAACCCUCGAAUGUCGGAACCACAAUCCGAUGUGUUAACCACUUCACCACAGCCGCCAUG---------GCAGGGGC .....((((.-----...........((((((((.((((.......)))).(((.((((((.......))))))(((.....))))))....))))))))...---------....)))) ( -37.31) >sp_mu.0 1839095 119 - 2030921/40-160 CUAUACCCCAAUAAUAUAAUAAUAUAGGCGAGUGAUGGGGAUCGAACCCACGCAUGCCAGAGCCACAAUCUGGUGUGUUAACCACUUCACCACACCCGCCAAAACA-UAUUAACACGGGC ......(((.....((((....))))((((.(((.((((.......)))).((((((((((.......))))))))))..............))).))))......-.........))). ( -34.40) >sp_pn.0 1850856 114 + 2038615/40-160 CUAUACCCCAAUA------UAAAAUAGGCGAGUGAUGGGGAUCGAACCCACGCAUGCCAGAGCCACAAUCUGGUGUGUUAACCACUUCACCACACCCGCCAUAAUUCUAUUAACACGGGC ......(((....------.......((((.(((.((((.......)))).((((((((((.......))))))))))..............))).)))).((((...))))....))). ( -33.10) >sp_ag.0 1904059 110 - 2160267/40-160 CUAUACCCCA---------UGAAA-AGGCGAGUGAUGGGAAUCGAACCCACGAAUGUCAGAGCCACAAUCUGAUGUGUUAACCACUUCACCACACCCGCCAUAUUAGAAAAAACACGGGC ......(((.---------(....-)((((.(((.(((((((((......)))((((((((.......)))))))).........))).)))))).))))................))). ( -28.10) >consensus CUAUACCCCA_____AUAAUAAUAAAGGCGAGUGAUGGGAAUCGAACCCACGAAUGUCAGAACCACAAUCCGAUGUGUUAACCACUUCACCACACCCGCCAUA_________ACACGGGC ..........................((((.(((.((((.......))))...((((((((.......))))))))................))).)))).................... (-26.83 = -24.78 + -2.05)

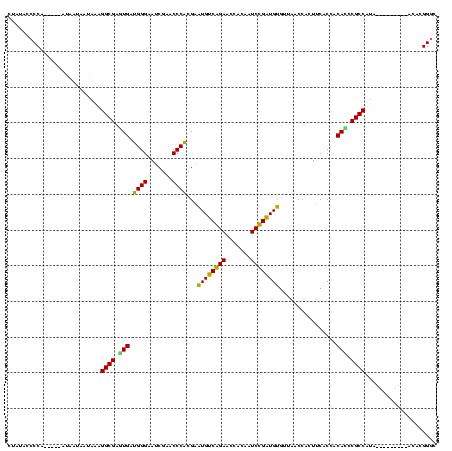
| Location | 1,574,809 – 1,574,918 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.43 |
| Mean single sequence MFE | -35.94 |
| Consensus MFE | -30.21 |
| Energy contribution | -28.27 |
| Covariance contribution | -1.94 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.03 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.757186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1574809 109 - 2685015/40-160 GUCCCUGC---------CAUGGCGGUUGUGGCGAAGUGGUUAACGCAUCGGAUUGUGGUUCCGACACUCGAGGGUUCGAUUCCCUUCAACCGCCCUUAAUUUUAU--UAAUGGGCUAUAG ......((---------(..((((((((...(((.(((((....)).(((((.......)))))))))))((((.......)))).)))))))).(((((...))--)))..)))..... ( -32.60) >sy_sa.0 1549924 106 - 2516575/40-160 GCCCCUGC---------CAUGGCGGUUGUGGCGAAGUGGUUAACGCAUCGGAUUGUGGUUCCGACACUCGUGGGUUCGAUUCCCAUCAUCCGCCCUUAGUAUUAU-----UGGGCUAUAG ((((((((---------....))))....(((((.(((.....))).))((((.((((...(((.(((....))))))....)))).)))))))...........-----.))))..... ( -34.90) >sy_au.0 1974979 106 + 2809422/40-160 GCCCCUGC---------CAUGGCGGCUGUGGUGAAGUGGUUAACACAUCGGAUUGUGGUUCCGACAUUCGAGGGUUCGAUCCCCUUCAGCCGCCCUUAUUAUUAA-----UGGGCUAUAG ((((....---------...((((((((.((.((.(((.....))).))((((((.((..(((.....)).)..))))))))))..)))))))).(((....)))-----.))))..... ( -38.90) >sp_mu.0 1839095 119 - 2030921/40-160 GCCCGUGUUAAUA-UGUUUUGGCGGGUGUGGUGAAGUGGUUAACACACCAGAUUGUGGCUCUGGCAUGCGUGGGUUCGAUCCCCAUCACUCGCCUAUAUUAUUAUAUUAUUGGGGUAUAG .((((...(((((-((....(((((((((((.(..(((.....)))....(((((.(((((..(....)..)))))))))))))).))))))))........))))))).))))...... ( -36.70) >sp_pn.0 1850856 114 + 2038615/40-160 GCCCGUGUUAAUAGAAUUAUGGCGGGUGUGGUGAAGUGGUUAACACACCAGAUUGUGGCUCUGGCAUGCGUGGGUUCGAUCCCCAUCACUCGCCUAUUUUA------UAUUGGGGUAUAG .((((.....(((((((...(((((((((((.(..(((.....)))....(((((.(((((..(....)..)))))))))))))).)))))))).))))))------)..))))...... ( -36.60) >sp_ag.0 1904059 110 - 2160267/40-160 GCCCGUGUUUUUUCUAAUAUGGCGGGUGUGGUGAAGUGGUUAACACAUCAGAUUGUGGCUCUGACAUUCGUGGGUUCGAUUCCCAUCACUCGCCU-UUUCA---------UGGGGUAUAG .((((((.............((((((((((((((.(((.....))).)))(((((.(((((((.....)).)))))))))).))).)))))))).-...))---------))))...... ( -35.93) >consensus GCCCCUGC_________CAUGGCGGGUGUGGUGAAGUGGUUAACACAUCAGAUUGUGGCUCCGACAUUCGUGGGUUCGAUCCCCAUCACCCGCCCUUAUUAUUAU_____UGGGCUAUAG .(((..............(.((((((((..(((..(((.....))).(((((.......))))))))..(((((.......))))))))))))).)...............)))...... (-30.21 = -28.27 + -1.94) # Strand winner: reverse (0.93)

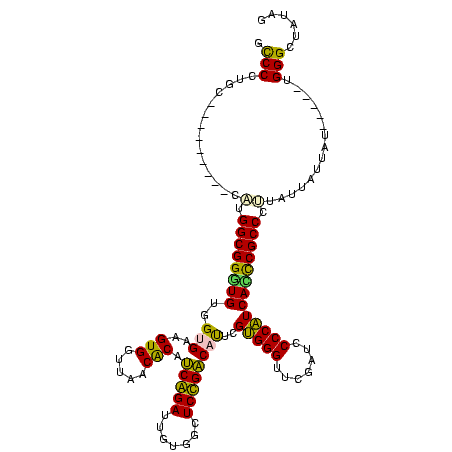
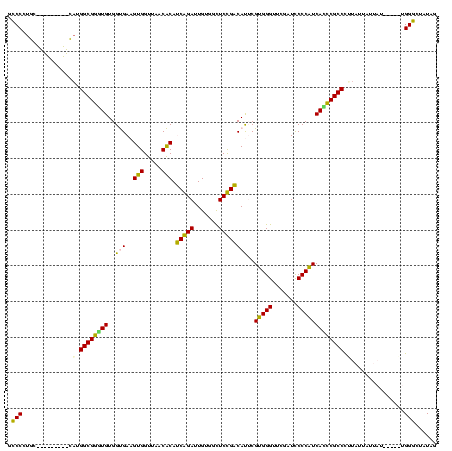
| Location | 1,574,809 – 1,574,914 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.94 |
| Mean single sequence MFE | -24.43 |
| Consensus MFE | -20.04 |
| Energy contribution | -19.18 |
| Covariance contribution | -0.86 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922611 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1574809 105 - 2685015/120-240 ACCACUUCGCCACAACCGCCAUG---------GCAGGGACAGUAGGAAUCGAACCCACAUCAAAGGUUUUGGAGACCUCUAUUCUACCGUUGAACUAUGUCCCUA---AAUUAAG---AU ........((((.........))---------))(((((((((.((........)))).((((.(((..((((....))))....))).))))....))))))).---.......---.. ( -26.50) >sy_sa.0 1549924 111 - 2516575/120-240 ACCACUUCGCCACAACCGCCAUG---------GCAGGGGCAGUAGGAAUCGAACCCACAUCAAAGGUUUUGGAGACCUCUAUUCUACCGUUGAAAUAUGCCCCUAUUAAAUUAAAUAAAU ........((((.........))---------))(((((((((.((........)))).((((.(((..((((....))))....))).))))....)))))))................ ( -28.60) >sy_au.0 1974979 108 + 2809422/120-240 ACCACUUCACCACAGCCGCCAUG---------GCAGGGGCAGUAGGAAUCGAACCCACACCAAAGGUUUUGGAGACCUCUAUUCUACCGUUGAACUAUGCCCCUAUUAAAAAUAAUA--- ..............(((.....)---------))(((((((((.((........))))..(((.(((..((((....))))....))).))).....))))))).............--- ( -26.60) >sp_mu.0 1839095 108 - 2030921/120-240 ACCACUUCACCACACCCGCCAAAACA-UAUUAACACGGGCAGUAGGAAUUGAACCCACACUGAAGGUUUUGGAGACCUUAGUUCUACCUUUAAACUAUGCCCGUA---AGGU-------- ........(((......(......).-.......(((((((.(((...(((((.....(((((.((((.....)))))))))......))))).)))))))))).---.)))-------- ( -22.20) >sp_pn.0 1850856 107 + 2038615/120-240 ACCACUUCACCACACCCGCCAUAAUUCUAUUAACACGGGCAGUAGGAAUUGAACCCACACUGAAGGUUUUGGAGACCUUAGUUCUACCUUUAAACUAUGCCCGUA---AA---------- ..................................(((((((.(((...(((((.....(((((.((((.....)))))))))......))))).)))))))))).---..---------- ( -21.90) >sp_ag.0 1904059 109 - 2160267/120-240 ACCACUUCACCACACCCGCCAUAUUAGAAAAAACACGGGCAGUAGGAAUCGAACCCACACUGAAGGUUUUGGAGACCUUAGUUCUACCUUUAAACUAUGCCCGUU---UACU-------- ..................................(((((((((.((........))))(((((.((((.....)))))))))...............))))))).---....-------- ( -20.80) >consensus ACCACUUCACCACACCCGCCAUA_________ACACGGGCAGUAGGAAUCGAACCCACACCAAAGGUUUUGGAGACCUCAAUUCUACCGUUAAACUAUGCCCCUA___AAUU________ ..................................(((((((((.((........))))......(((((.(((((.......))).))...))))).)))))))................ (-20.04 = -19.18 + -0.86)

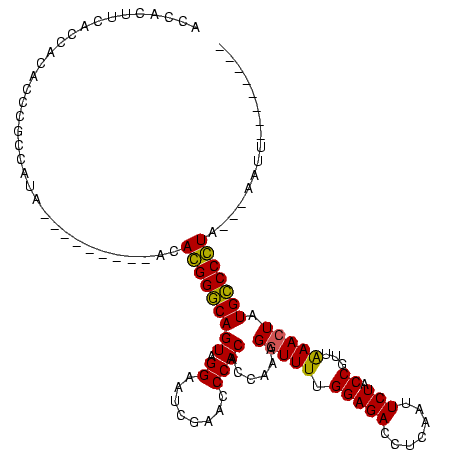
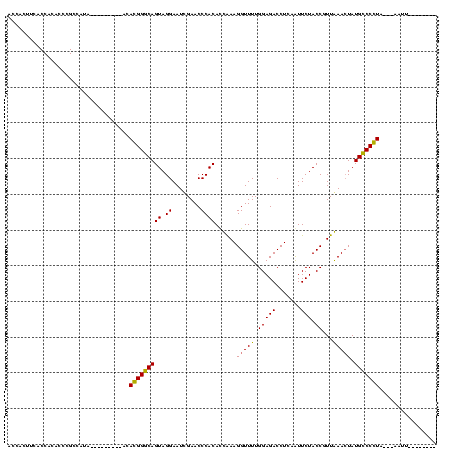
| Location | 1,574,809 – 1,574,927 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.09 |
| Mean single sequence MFE | -46.30 |
| Consensus MFE | -45.51 |
| Energy contribution | -45.73 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.635406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1574809 118 - 2685015/240-360 GGUGGAGGGGGGCAGAUUCGAACUGCCGAACCCGAAGGAGCGGAUUUACAGUCCGCCGCGUUUAGCCACUUCGCUACCCCUCCAUGA--GAUGGUGCCGGCCAGAGGACUUGAACCCCCA .((((((((((((((.......))))).....((((((.((((((.....)))))))((.....))..)))))...)))))))))..--..((((....))))(.((........)).). ( -46.70) >sy_sa.0 1549924 120 - 2516575/240-360 GGUGGAGGGGGGCAGAUUCGAACUGCCGAACCCGAAGGAGCGGAUUUACAGUCCGCCGCGUUUAGCCACUUCGCUACCCCUCCAAAAAAGAUGGUGCCGGCCAGAGGACUUGAACCCCCA (((((((((((((((.......))))).....((((((.((((((.....)))))))((.....))..)))))...))))))))........(((.(.((((...)).)).).))).)). ( -45.90) >sy_au.0 1974979 117 + 2809422/240-360 AAUGGAGGGGGGCAGAUUCGAACUGCCGAACCCGAAGGAGCGGAUUUACAGUCCGCCGCGUUUAGCCACUUCGCUACCCCUCCAUAA---AUGGUGCCGGCCAGAGGACUUGAACCCCCA .((((((((((((((.......))))).....((((((.((((((.....)))))))((.....))..)))))...)))))))))..---.((((....))))(.((........)).). ( -46.30) >consensus GGUGGAGGGGGGCAGAUUCGAACUGCCGAACCCGAAGGAGCGGAUUUACAGUCCGCCGCGUUUAGCCACUUCGCUACCCCUCCAUAA__GAUGGUGCCGGCCAGAGGACUUGAACCCCCA .((((((((((((((.......))))).....((((((.((((((.....)))))))((.....))..)))))...)))))))))......((((....))))(.((........)).). (-45.51 = -45.73 + 0.22)

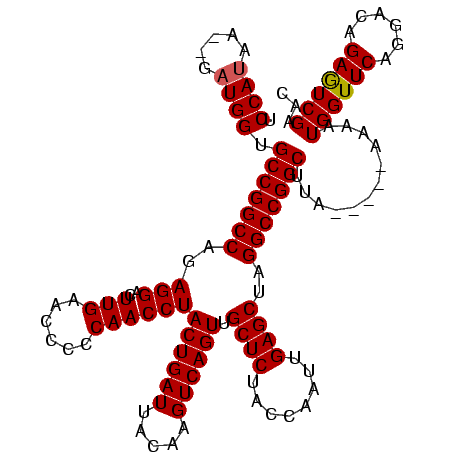
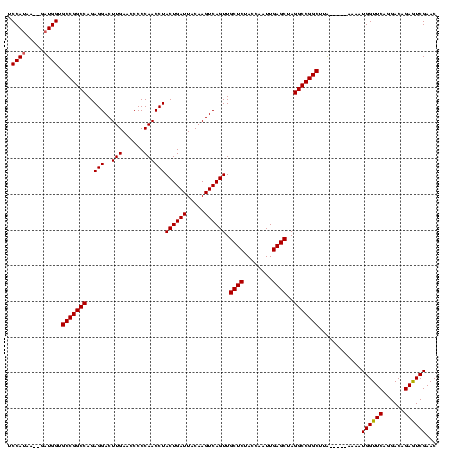
| Location | 1,574,809 – 1,574,924 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.42 |
| Mean single sequence MFE | -35.53 |
| Consensus MFE | -33.32 |
| Energy contribution | -33.43 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.859617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1574809 115 - 2685015/320-440 UCCAUGA--GAUGGUGCCGGCCAGAGGACUUGAACCCCCAACCUACUGAUUACAAGUCAGUUGCUCUACCAAUUGAGCUAGGCCGGCAUAAC---AAAAUGGUUCAGGACAGAGUCGAAC .((((..--....(((((((((..(((..(((......))))))((((((.....)))))).((((........))))..)))))))))...---...)))).....(((...))).... ( -37.70) >sy_sa.0 1549924 120 - 2516575/320-440 UCCAAAAAAGAUGGUGCCGGCCAGAGGACUUGAACCCCCAACCUACUGAUUACAAGUCAGUUGCUCUACCAGUUGAGCUAGGCCGGCUUGCAUUAAAAAUGGUUCAGGACAGAAUCGAAC .........((((..(((((((..(((..(((......))))))((((((.....)))))).((((........))))..)))))))...)))).....((((((......))))))... ( -36.00) >sy_au.0 1974979 112 + 2809422/320-440 UCCAUAA---AUGGUGCCGGCCAGAGGACUUGAACCCCCAACCUACUGAUUACAAGUCAGUUGCUCUACCAAUUGAGCUAGGCCGGCUAA-----GAAAUGGUUCAGGACAGAGUCGAAC .(((...---.))).(((((((..(((..(((......))))))((((((.....)))))).((((........))))..)))))))...-----....((..((......))..))... ( -32.90) >consensus UCCAUAA__GAUGGUGCCGGCCAGAGGACUUGAACCCCCAACCUACUGAUUACAAGUCAGUUGCUCUACCAAUUGAGCUAGGCCGGCUUA_____AAAAUGGUUCAGGACAGAGUCGAAC .((((.....)))).(((((((..(((..(((......))))))((((((.....)))))).((((........))))..)))))))............((((((......))))))... (-33.32 = -33.43 + 0.11)

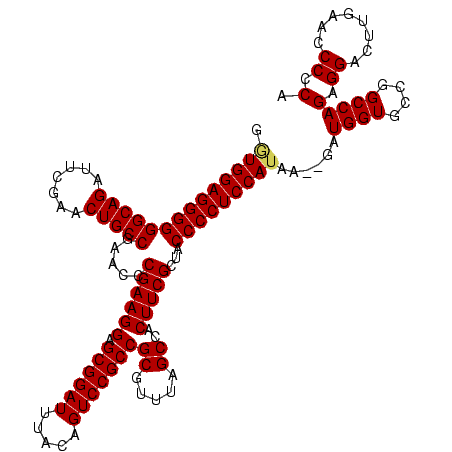
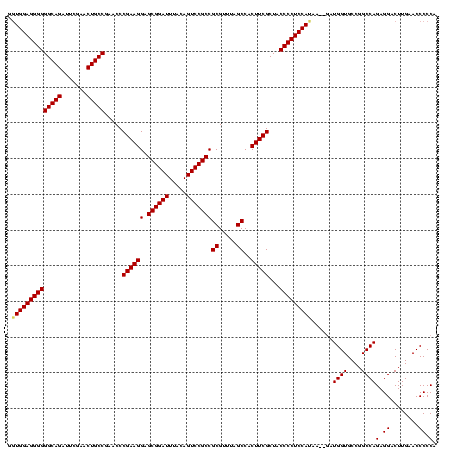
| Location | 1,574,809 – 1,574,919 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.32 |
| Mean single sequence MFE | -32.68 |
| Consensus MFE | -24.90 |
| Energy contribution | -26.15 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.76 |
| SVM decision value | 3.24 |
| SVM RNA-class probability | 0.998829 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1574809 110 - 2685015/400-520 GGCCGGCAUAAC---AAAAUGGUUCAGGACAGAGUCGAACUGCCGACACAUGGAGCUUCAAUCCAUUGCUCUACCAACUGAGCUACUGAACCAUUAUUUG-------UAAUGGCGGUCUC (((((.(((.((---((((((((((((......((((......))))..(((((.......))))).((((........))))..)))))))))..))))-------).))).))))).. ( -44.30) >sy_sa.0 1549924 110 - 2516575/400-520 GGCCGGCUUGCAUUAAAAAUGGUUCAGGACAGAAUCGAACUGCCGACACAUGGAGCUUCAAUCCAUUGCUCUACCAACUGAGCUACUGAACCAUAAUA----------AAUGGCGGUCCC (((((.(....((((....((((((((.......(((......)))...(((((.......))))).((((........))))..)))))))))))).----------...).))))).. ( -31.60) >sy_au.0 1974979 115 + 2809422/400-520 GGCCGGCUAA-----GAAAUGGUUCAGGACAGAGUCGAACUGCCGACACAUGGAGCUUCAAUCCAUUGCUCUACCAACUGAGCUACUGAACCAUAAUAAAAAUGUAAUGAUGGCGGUCUC (((((.(((.-----...(((((((((......((((......))))..(((((.......))))).((((........))))..))))))))).(((....))).....)))))))).. ( -35.20) >sp_mu.0 1839095 84 - 2030921/400-520 ---------------AUGAUGGCGCGAGACGGAAUCGAACCGCCGACACAUGGAGCUUCAAUCCAUUGCUCUACCAACUGAGCUACCGAGCCUUCCUCU--------------------- ---------------..((.(((.((.(.(((.......))).).....(((((.......))))).((((........))))...)).))).))....--------------------- ( -19.60) >consensus GGCCGGCUUA_____AAAAUGGUUCAGGACAGAAUCGAACUGCCGACACAUGGAGCUUCAAUCCAUUGCUCUACCAACUGAGCUACUGAACCAUAAUAU_________AAUGGCGGUCUC (((((.(..........((((((((((....(.((((......)))).)(((((.......))))).((((........))))..))))))))))................).))))).. (-24.90 = -26.15 + 1.25)

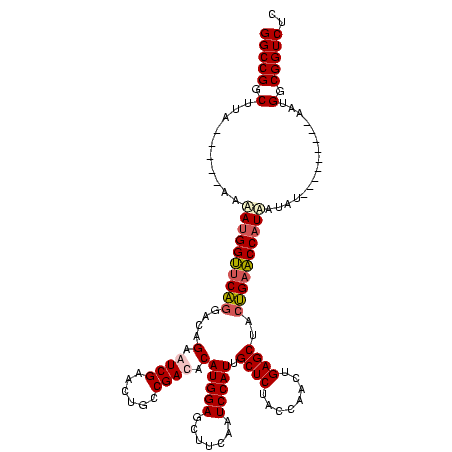
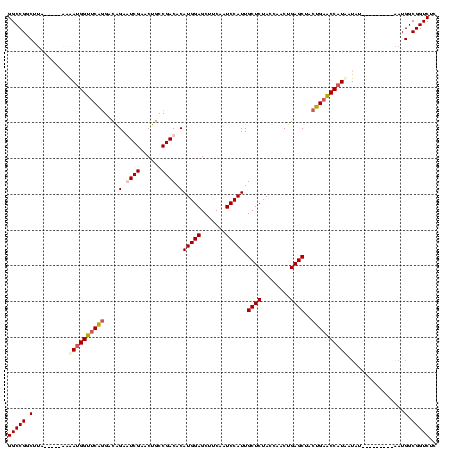
| Location | 1,574,809 – 1,574,919 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.32 |
| Mean single sequence MFE | -31.65 |
| Consensus MFE | -27.00 |
| Energy contribution | -26.75 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.19 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.791021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1574809 110 - 2685015/400-520 GAGACCGCCAUUA-------CAAAUAAUGGUUCAGUAGCUCAGUUGGUAGAGCAAUGGAUUGAAGCUCCAUGUGUCGGCAGUUCGACUCUGUCCUGAACCAUUUU---GUUAUGCCGGCC ....(((.(((.(-------((((..(((((((((..((((........)))).(((((.......))))).....(((((.......)))))))))))))))))---)).))).))).. ( -36.40) >sy_sa.0 1549924 110 - 2516575/400-520 GGGACCGCCAUU----------UAUUAUGGUUCAGUAGCUCAGUUGGUAGAGCAAUGGAUUGAAGCUCCAUGUGUCGGCAGUUCGAUUCUGUCCUGAACCAUUUUUAAUGCAAGCCGGCC .((.((((((((----------....(((((((((..((((........))))...((((((((.((((.......)).))))))))))....)))))))))....))))...).))))) ( -34.10) >sy_au.0 1974979 115 + 2809422/400-520 GAGACCGCCAUCAUUACAUUUUUAUUAUGGUUCAGUAGCUCAGUUGGUAGAGCAAUGGAUUGAAGCUCCAUGUGUCGGCAGUUCGACUCUGUCCUGAACCAUUUC-----UUAGCCGGCC ......(((.....((......))..(((((((((..((((........)))).(((((.......))))).....(((((.......))))))))))))))...-----......))). ( -27.90) >sp_mu.0 1839095 84 - 2030921/400-520 ---------------------AGAGGAAGGCUCGGUAGCUCAGUUGGUAGAGCAAUGGAUUGAAGCUCCAUGUGUCGGCGGUUCGAUUCCGUCUCGCGCCAUCAU--------------- ---------------------....((.(((..((.(((((((((.((......)).))))).))))))..(((..(((((.......))))).)))))).))..--------------- ( -28.20) >consensus GAGACCGCCAUU_________AUAUUAUGGUUCAGUAGCUCAGUUGGUAGAGCAAUGGAUUGAAGCUCCAUGUGUCGGCAGUUCGACUCUGUCCUGAACCAUUUU_____UAAGCCGGCC ......(((................((((((((((..((((........)))).(((((.......))))).....(((((.......))))))))))))))).............))). (-27.00 = -26.75 + -0.25) # Strand winner: reverse (0.82)

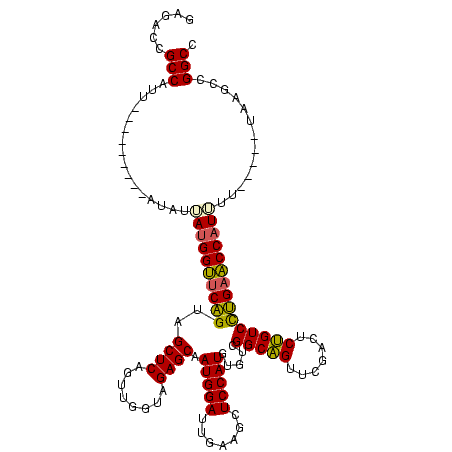
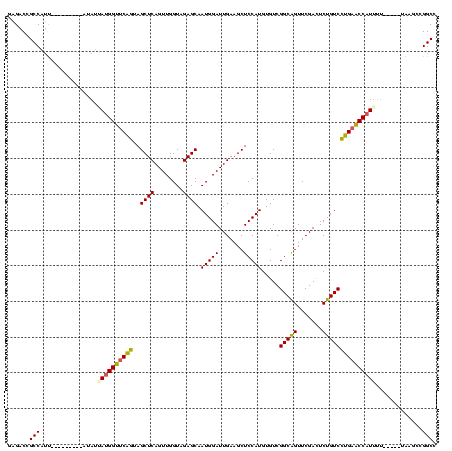
| Location | 1,574,809 – 1,574,922 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.19 |
| Mean single sequence MFE | -32.47 |
| Consensus MFE | -30.60 |
| Energy contribution | -29.71 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1574809 113 - 2685015/480-600 AGCUACUGAACCAUUAUUUG-------UAAUGGCGGUCUCGACGGGAAUCGAACCCGCGAUCUCCUGCGUGACAGGCAGGCGUGUUAACCGCUACACUACGAGACCUAAAAGAUAAAUUG ..........((((((....-------)))))).(((((((..((..((((......))))..)).((((((((.(....).)))))..))).......))))))).............. ( -33.70) >sy_sa.0 1549924 108 - 2516575/480-600 AGCUACUGAACCAUAAUA----------AAUGGCGGUCCCGAUGGGAUUCGAACCCACGAUCUCCUGCGUGACAGGCAGGCGUGUUAACCGCUACACUACGGGACCUUUUAAA--AAUUG ..........((((....----------.)))).(((((((.((((.......))))......(((((.......))))).((((........))))..))))))).......--..... ( -31.80) >sy_au.0 1974979 118 + 2809422/480-600 AGCUACUGAACCAUAAUAAAAAUGUAAUGAUGGCGGUCUCGACGGGAAUCGAACCCGCGAUCUCCUGCGUGACAGGCAGGCGUGUUAACCGCUACACUACGAGACCUAUAAAA--UAUUG .((((......(((.......)))......))))(((((((..((..((((......))))..)).((((((((.(....).)))))..))).......))))))).......--..... ( -31.90) >consensus AGCUACUGAACCAUAAUA_________UAAUGGCGGUCUCGACGGGAAUCGAACCCGCGAUCUCCUGCGUGACAGGCAGGCGUGUUAACCGCUACACUACGAGACCUAUAAAA__AAUUG .((((.........................))))(((((((.((((.......))))......(((((.......))))).((((........))))..))))))).............. (-30.60 = -29.71 + -0.89)

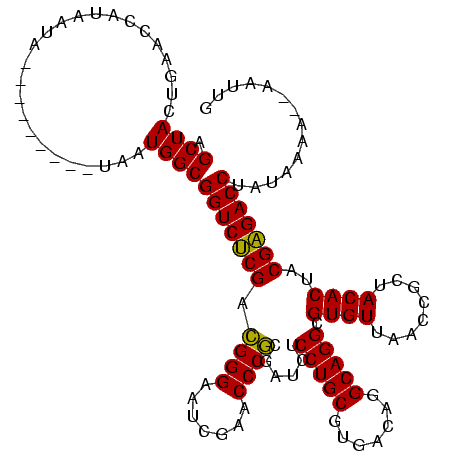
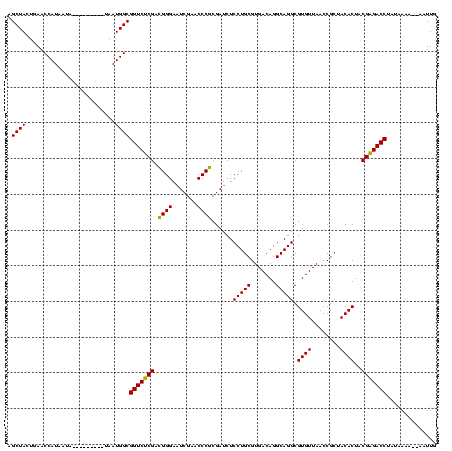
| Location | 1,574,809 – 1,574,922 |
|---|---|
| Length | 113 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.19 |
| Mean single sequence MFE | -34.83 |
| Consensus MFE | -33.35 |
| Energy contribution | -32.46 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1574809 113 - 2685015/480-600 CAAUUUAUCUUUUAGGUCUCGUAGUGUAGCGGUUAACACGCCUGCCUGUCACGCAGGAGAUCGCGGGUUCGAUUCCCGUCGAGACCGCCAUUA-------CAAAUAAUGGUUCAGUAGCU ..............(((((((.......((((((......(((((.......))))).))))))(((.......)))..)))))))(((((((-------....)))))))......... ( -36.10) >sy_sa.0 1549924 108 - 2516575/480-600 CAAUU--UUUAAAAGGUCCCGUAGUGUAGCGGUUAACACGCCUGCCUGUCACGCAGGAGAUCGUGGGUUCGAAUCCCAUCGGGACCGCCAUU----------UAUUAUGGUUCAGUAGCU .....--.......(((((((..((((........)))).(((((.......))))).....(((((.......))))))))))))(((((.----------....)))))......... ( -34.70) >sy_au.0 1974979 118 + 2809422/480-600 CAAUA--UUUUAUAGGUCUCGUAGUGUAGCGGUUAACACGCCUGCCUGUCACGCAGGAGAUCGCGGGUUCGAUUCCCGUCGAGACCGCCAUCAUUACAUUUUUAUUAUGGUUCAGUAGCU .....--.............((((((..((((((....(.(((((.......))))).).(((((((.......)))).)))))))))...))))))....................... ( -33.70) >consensus CAAUU__UUUUAUAGGUCUCGUAGUGUAGCGGUUAACACGCCUGCCUGUCACGCAGGAGAUCGCGGGUUCGAUUCCCGUCGAGACCGCCAUUA_________UAUUAUGGUUCAGUAGCU ..............(((((((..((((........)))).(((((.......))))).....(((((.......))))))))))))(((((...............)))))......... (-33.35 = -32.46 + -0.89) # Strand winner: reverse (0.93)

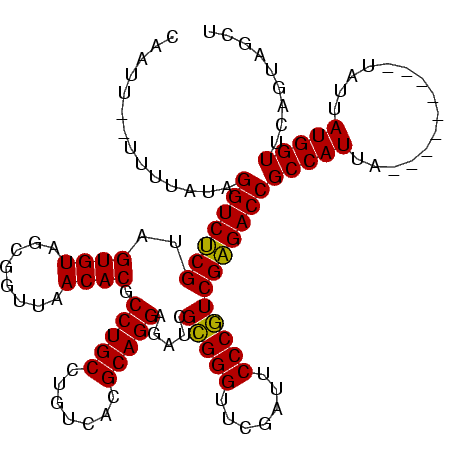
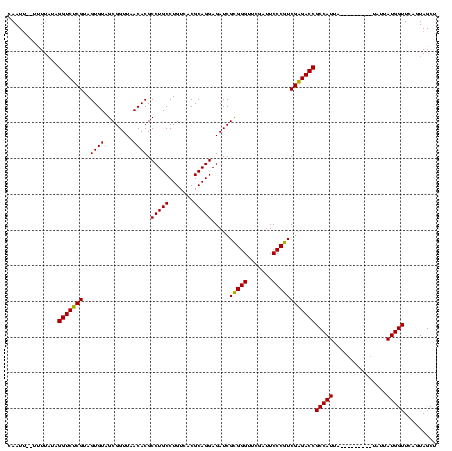
| Location | 1,574,809 – 1,574,929 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.85 |
| Mean single sequence MFE | -36.47 |
| Consensus MFE | -35.03 |
| Energy contribution | -35.37 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.715014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1574809 120 - 2685015/560-680 CGUGUUAACCGCUACACUACGAGACCUAAAAGAUAAAUUGCGGGAGGCGGAUUUGAACCACCGACCUUCGGGUUAUGAGCCCGACGAGCUACCGAACUGCUCCAUCCCGCGAUAAUAAUU .((((........))))...................((((((((((((((..........))).)).((((((.....)))))).((((.........))))..)))))))))....... ( -36.20) >sy_sa.0 1549924 118 - 2516575/560-680 CGUGUUAACCGCUACACUACGGGACCUUUUAAA--AAUUGCGGGAGGCGGAUUUGAACCACCGACCUUCGGGUUAUGAGCCCGACGAGCUACCGAACUGCUCCAUCCCGCGUUAAUAAUA ..(((((((.......(....)...........--....(((((((((((..........))).)).((((((.....)))))).((((.........))))..)))))))))))))... ( -35.60) >sy_au.0 1974979 118 + 2809422/560-680 CGUGUUAACCGCUACACUACGAGACCUAUAAAA--UAUUGCGGGAGGCGGAUUUGAACCACCGACCUUCGGGUUAUGAGCCCGACGAGCUACCGAACUGCUCCAUCCCGCGAUAAUAAAA .((((........))))................--(((((((((((((((..........))).)).((((((.....)))))).((((.........))))..))))))))))...... ( -37.60) >consensus CGUGUUAACCGCUACACUACGAGACCUAUAAAA__AAUUGCGGGAGGCGGAUUUGAACCACCGACCUUCGGGUUAUGAGCCCGACGAGCUACCGAACUGCUCCAUCCCGCGAUAAUAAUA .((((........))))...................((((((((((((((..........))).)).((((((.....)))))).((((.........))))..)))))))))....... (-35.03 = -35.37 + 0.33)

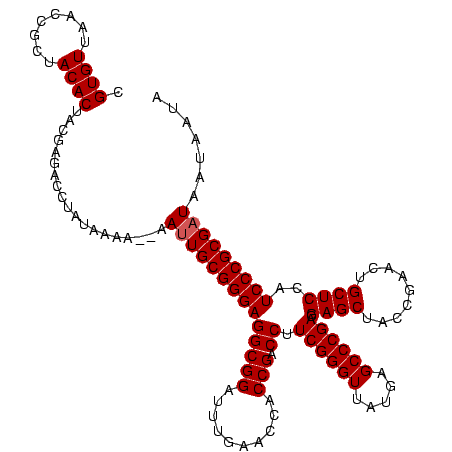
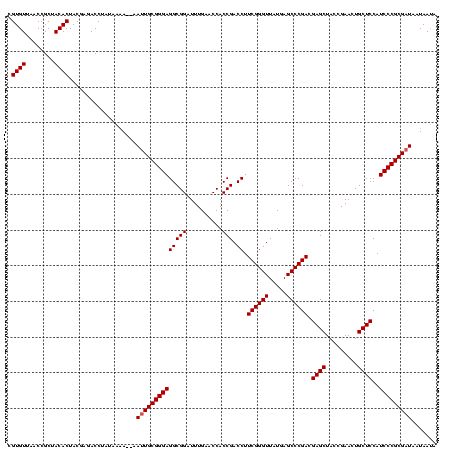
| Location | 1,574,809 – 1,574,920 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.77 |
| Mean single sequence MFE | -34.83 |
| Consensus MFE | -29.00 |
| Energy contribution | -26.95 |
| Covariance contribution | -2.05 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.936706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1574809 111 - 2685015/680-800 ---AAUGGCGGAGGAAGAGGGAUUCGAACCCCCGCGGCCCGUUAAGGCCCUGUCGGUUUUCAAGACCGAUCCCUUCAGCCGGACUUGGGUAUUCCUCCACUAAU------UAAAUGUAAA ---..(((.(((((((((((((..........((.((((......)))).))((((((.....))))))))))))..(((.......))).))))))).)))..------.......... ( -38.30) >sy_sa.0 1549924 119 - 2516575/680-800 UA-AAUGGCGGAGGAAGAGGGAUUCGAACCCCCGCGAGCCGUUAAGCUCCUGUCGGUUUUCAAGACCGAUCCCUUCAGCCGGACUUGGGUAUUCCUCCACAAAUAUUAGCUAAAAAUAAA ..-..(((((((((((((((((..........((.((((......)))).))((((((.....))))))))))))..(((.......))).)))))))..........))))........ ( -34.30) >sy_au.0 1974979 119 + 2809422/680-800 AAUAAUGGCGGAGGAAGAGGGAUUCGAACCCCCGCGGCCCGUUAAGGCCCUGUCGGUUUUCAAGACCGAUCCCUUCAGCCGGACUUGGGUAUUCCUCCAUUAUUAU-AGGUAAAUCGCUA .(((((((.(((((((((((((..........((.((((......)))).))((((((.....))))))))))))..(((.......))).))))))).)))))))-............. ( -39.50) >sp_mu.0 1839095 111 - 2030921/680-800 -----UAAAGGAGGAUGUGGGAUUCGAACCCACGCACGCUUUUACACGCCUGACGGUUUUCAAGACCGUUCCCUUCAGCCGGACUUGGGUAAUCCUCCAAUAAAUA----UAAUAAUAGU -----....((((((((((((.......)))))....((((.....((.(((((((((.....)))))......)))).)).....)))).)))))))........----.......... ( -32.30) >sp_pn.0 1850856 101 + 2038615/680-800 ------AAAGGAGGAUGUGGGAUUCGAACCCACGCACGCUUUUACACGCCUGACGGUUUUCAAGACCGUUCCCUUCAGCCGGACUUGGGUAAUCCUCCAA-----------AAUAGU--U ------...((((((((((((.......)))))....((((.....((.(((((((((.....)))))......)))).)).....)))).)))))))..-----------......--. ( -32.30) >sp_ag.0 1904059 110 - 2160267/680-800 -----UAAAGGAGGAUGUGGGAUUCGAACCCACGCACGCUUUUACACGCCUGACGGUUUUCAAGACCGUUCCCUUCAGCCGGACUUGGGUAAUCCUCCAUAUAACA-----AAAAAUAGU -----....((((((((((((.......)))))....((((.....((.(((((((((.....)))))......)))).)).....)))).)))))))........-----......... ( -32.30) >consensus _____UAAAGGAGGAAGAGGGAUUCGAACCCACGCACGCCGUUAAACGCCUGACGGUUUUCAAGACCGAUCCCUUCAGCCGGACUUGGGUAAUCCUCCAAUAAU______UAAAAAUAAA .........(((((((..((((..........((..((........))..))((((((.....))))))))))....(((.......))).)))))))...................... (-29.00 = -26.95 + -2.05)
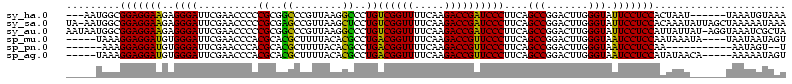
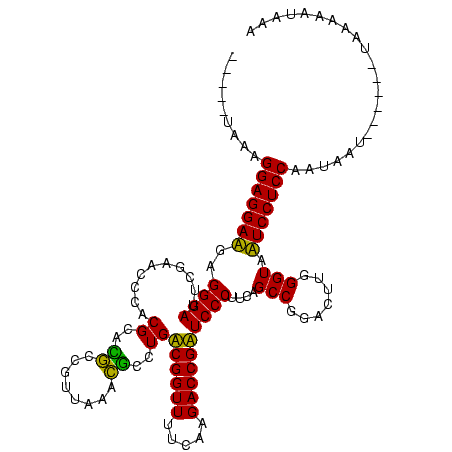
| Location | 1,574,809 – 1,574,920 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.77 |
| Mean single sequence MFE | -35.62 |
| Consensus MFE | -36.15 |
| Energy contribution | -33.10 |
| Covariance contribution | -3.05 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.37 |
| Structure conservation index | 1.02 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1574809 111 - 2685015/680-800 UUUACAUUUA------AUUAGUGGAGGAAUACCCAAGUCCGGCUGAAGGGAUCGGUCUUGAAAACCGACAGGGCCUUAACGGGCCGCGGGGGUUCGAAUCCCUCUUCCUCCGCCAUU--- ..........------....(((((((((....((((.((((((....)).)))).))))...........(((((....)))))..(((((.......))))))))))))))....--- ( -40.00) >sy_sa.0 1549924 119 - 2516575/680-800 UUUAUUUUUAGCUAAUAUUUGUGGAGGAAUACCCAAGUCCGGCUGAAGGGAUCGGUCUUGAAAACCGACAGGAGCUUAACGGCUCGCGGGGGUUCGAAUCCCUCUUCCUCCGCCAUU-UA ....................(((((((((....((((.((((((....)).)))).))))...........(((((....)))))..(((((.......))))))))))))))....-.. ( -37.70) >sy_au.0 1974979 119 + 2809422/680-800 UAGCGAUUUACCU-AUAAUAAUGGAGGAAUACCCAAGUCCGGCUGAAGGGAUCGGUCUUGAAAACCGACAGGGCCUUAACGGGCCGCGGGGGUUCGAAUCCCUCUUCCUCCGCCAUUAUU .............-(((((..((((((((....((((.((((((....)).)))).))))...........(((((....)))))..(((((.......)))))))))))))..))))). ( -38.20) >sp_mu.0 1839095 111 - 2030921/680-800 ACUAUUAUUA----UAUUUAUUGGAGGAUUACCCAAGUCCGGCUGAAGGGAACGGUCUUGAAAACCGUCAGGCGUGUAAAAGCGUGCGUGGGUUCGAAUCCCACAUCCUCCUUUA----- ..........----........(((((((....((((.(((...........))).))))......(.((.((........)).)))(((((.......))))))))))))....----- ( -32.60) >sp_pn.0 1850856 101 + 2038615/680-800 A--ACUAUU-----------UUGGAGGAUUACCCAAGUCCGGCUGAAGGGAACGGUCUUGAAAACCGUCAGGCGUGUAAAAGCGUGCGUGGGUUCGAAUCCCACAUCCUCCUUU------ .--......-----------..(((((((....((((.(((...........))).))))......(.((.((........)).)))(((((.......))))))))))))...------ ( -32.60) >sp_ag.0 1904059 110 - 2160267/680-800 ACUAUUUUU-----UGUUAUAUGGAGGAUUACCCAAGUCCGGCUGAAGGGAACGGUCUUGAAAACCGUCAGGCGUGUAAAAGCGUGCGUGGGUUCGAAUCCCACAUCCUCCUUUA----- .........-----........(((((((....((((.(((...........))).))))......(.((.((........)).)))(((((.......))))))))))))....----- ( -32.60) >consensus ACUAUUUUUA______AUUAAUGGAGGAAUACCCAAGUCCGGCUGAAGGGAACGGUCUUGAAAACCGACAGGCGCGUAAAAGCGCGCGGGGGUUCGAAUCCCACAUCCUCCGCCA_____ ......................(((((((..(((.((.....))...))).(((((.......)))))...((((......))))..(((((.......))))))))))))......... (-36.15 = -33.10 + -3.05) # Strand winner: reverse (0.99)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:54:06 2006