| Sequence ID | sy_sa.0 |
|---|---|
| Location | 1,571,538 – 1,571,653 |
| Length | 115 |
| Max. P | 0.999938 |

| Location | 1,571,538 – 1,571,653 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.62 |
| Mean single sequence MFE | -29.80 |
| Consensus MFE | -21.42 |
| Energy contribution | -20.77 |
| Covariance contribution | -0.65 |
| Combinations/Pair | 1.32 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.72 |
| SVM decision value | 4.20 |
| SVM RNA-class probability | 0.999835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 1571538 115 + 2516575/0-120 UUGACGGCUAU-GAAGCCAAUACACACACAGAAAGUUAAA--GUAUAAAGGGUGCGAU--AUUUAAUUGUCGUGUUUAUAUGAGCUUUUUGUGGAGGUAAUAAACCAAUAGGAGGAAUUU .....((((..-..))))..(((...(((((((((((...--((((((...(..((((--(......)))))..))))))).)))))))))))...))).....((....))........ ( -32.40) >sy_ha.0 1712300 115 + 2685015/0-120 UUUUCGGCUAUAAAAGCCAAUACACACAUGAUAAACGUAC--AUUUAAAGGGUGCAAUUAAAUUAAUUGUC---UUUAUAUGAGUUUAUUAUGGAGGUAAAUAACCACUAGGAGGAAUUU (((((((((.....))))..(((...((((((((((...(--((.(((((...(((((((...))))))))---)))).))).))))))))))...)))............))))).... ( -28.30) >sy_au.0 1524840 117 - 2809422/0-120 UUUUCGGCUAU-AAAGCCAAUACACACAUAUCAAACGUGAUUGUGUAAAGGGUGCAAU--AUUUAUAUAUUGUCUUUACAAUAGUUUGCUAUGGAGGUAAUUAACCAAUAGGAGGAAUUU (((((((((..-..))))..(((...((((.(((((.......(((((((...(((((--((....))))))))))))))...))))).))))...)))............))))).... ( -28.70) >consensus UUUUCGGCUAU_AAAGCCAAUACACACAUAAAAAACGUAA__GUAUAAAGGGUGCAAU__AUUUAAUUGUCGU_UUUAUAUGAGUUUAUUAUGGAGGUAAUUAACCAAUAGGAGGAAUUU .....((((.....))))..(((...((((((((((......(((((((....(((((.......)))))....)))))))..))))))))))...))).....((....))........ (-21.42 = -20.77 + -0.65) # Strand winner: reverse (1.00)


| Location | 1,571,538 – 1,571,653 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.39 |
| Mean single sequence MFE | -19.16 |
| Consensus MFE | -9.87 |
| Energy contribution | -9.10 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.822164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 1571538 115 + 2516575/22-142 ACCUCCACAAAAAGCUCAUAUAAACACGACAAUUAAAU--AUCGCACCCUUUAUAC--UUUAACUUUCUGUGUGUGUAUUGGCUUC-AUAGCCGUCAAUAAAUAUAUCAUAUUUACCUAU ...........................(((.....(((--(.(((((.(.......--...........).)))))))))((((..-..)))))))..(((((((...)))))))..... ( -15.17) >sy_ha.0 1712300 115 + 2685015/22-142 ACCUCCAUAAUAAACUCAUAUAAA---GACAAUUAAUUUAAUUGCACCCUUUAAAU--GUACGUUUAUCAUGUGUGUAUUGGCUUUUAUAGCCGAAAACAAUAAUAUCAUAUUUCUAUAU .....(((.((((((.(((.((((---(.((((((...))))))....))))).))--)...)))))).)))(((...((((((.....))))))..))).................... ( -19.20) >sy_au.0 1524840 117 - 2809422/22-142 ACCUCCAUAGCAAACUAUUGUAAAGACAAUAUAUAAAU--AUUGCACCCUUUACACAAUCACGUUUGAUAUGUGUGUAUUGGCUUU-AUAGCCGAAAAUAAAUAUAACAUAAUGCCGUGU .....((..(((...((((((....)))))).....((--(((........((((((((((....)))).))))))..((((((..-..)))))).....))))).......)))..)). ( -23.10) >consensus ACCUCCAUAAAAAACUCAUAUAAA_ACGACAAUUAAAU__AUUGCACCCUUUAAAC__UUACGUUUAAUAUGUGUGUAUUGGCUUU_AUAGCCGAAAAUAAAUAUAUCAUAUUUCCAUAU .....((((.(((((...............................................))))).))))(((...((((((.....))))))..))).................... ( -9.87 = -9.10 + -0.77)

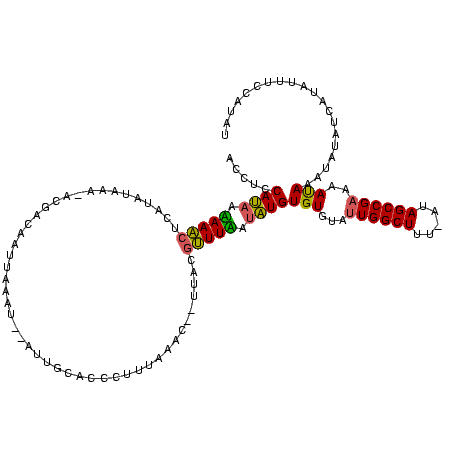
| Location | 1,571,538 – 1,571,653 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.39 |
| Mean single sequence MFE | -27.87 |
| Consensus MFE | -18.42 |
| Energy contribution | -17.77 |
| Covariance contribution | -0.65 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.66 |
| SVM decision value | 4.68 |
| SVM RNA-class probability | 0.999938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 1571538 115 + 2516575/22-142 AUAGGUAAAUAUGAUAUAUUUAUUGACGGCUAU-GAAGCCAAUACACACACAGAAAGUUAAA--GUAUAAAGGGUGCGAU--AUUUAAUUGUCGUGUUUAUAUGAGCUUUUUGUGGAGGU ...(((((((((...)))))))))...((((..-..))))...((...(((((((((((...--((((((...(..((((--(......)))))..))))))).)))))))))))...)) ( -32.20) >sy_ha.0 1712300 115 + 2685015/22-142 AUAUAGAAAUAUGAUAUUAUUGUUUUCGGCUAUAAAAGCCAAUACACACAUGAUAAACGUAC--AUUUAAAGGGUGCAAUUAAAUUAAUUGUC---UUUAUAUGAGUUUAUUAUGGAGGU .((((....))))..........((((((((.....))))........((((((((((...(--((.(((((...(((((((...))))))))---)))).))).)))))))))))))). ( -26.00) >sy_au.0 1524840 117 - 2809422/22-142 ACACGGCAUUAUGUUAUAUUUAUUUUCGGCUAU-AAAGCCAAUACACACAUAUCAAACGUGAUUGUGUAAAGGGUGCAAU--AUUUAUAUAUUGUCUUUACAAUAGUUUGCUAUGGAGGU .((..(((...((((............((((..-..))))..((((((...((((....)))))))))).((((.(((((--((....)))))))))))..))))...)))..))..... ( -25.40) >consensus AUACGGAAAUAUGAUAUAUUUAUUUUCGGCUAU_AAAGCCAAUACACACAUAAAAAACGUAA__GUAUAAAGGGUGCAAU__AUUUAAUUGUCGU_UUUAUAUGAGUUUAUUAUGGAGGU ...........................((((.....))))...((...((((((((((......(((((((....(((((.......)))))....)))))))..))))))))))...)) (-18.42 = -17.77 + -0.65) # Strand winner: reverse (1.00)

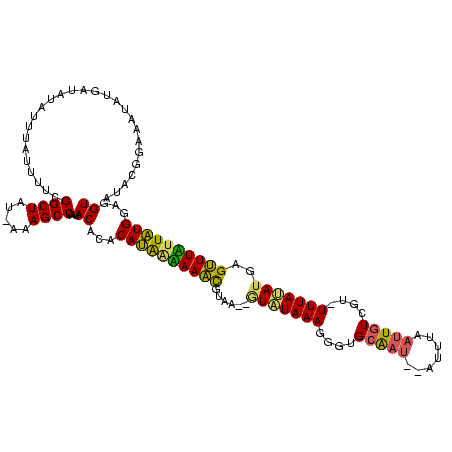
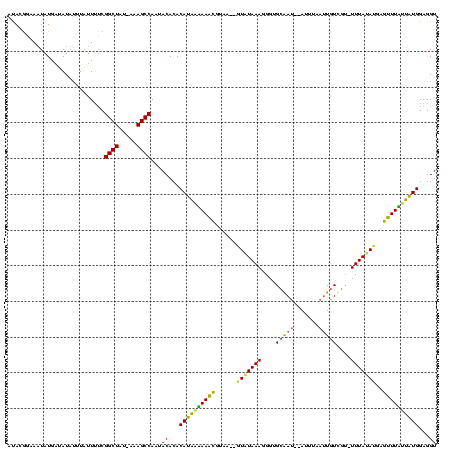
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:53:56 2006