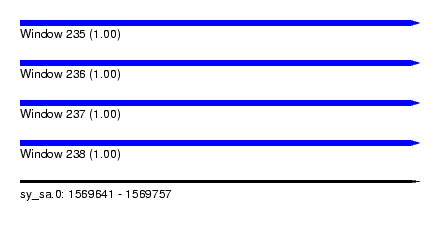
| Sequence ID | sy_sa.0 |
|---|---|
| Location | 1,569,641 – 1,569,757 |
| Length | 116 |
| Max. P | 0.999972 |

| Location | 1,569,641 – 1,569,757 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.37 |
| Mean single sequence MFE | -28.38 |
| Consensus MFE | -16.96 |
| Energy contribution | -17.86 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.96 |
| Structure conservation index | 0.60 |
| SVM decision value | 4.88 |
| SVM RNA-class probability | 0.999959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 1569641 116 + 2516575 UCUUAUCCUCUCUAUUGUAAAAAUAUGUAAAUUAUACAAGUAAAGAAGACACAAUAG-GUCUUCCAAUACAAAAGUGAAUCUUUCAU--ACGGAGCACAAAAGGUGUCUUCUUCCUUGU ...................................(((((..((((((((((.....-((.((((......((((.....))))...--..)))).)).....)))))))))).))))) ( -23.50) >sy_au.0 1522863 118 - 2809422 UCUUAUCCUCUCUAUCGUGAAUAUAUGUAACAUAUACAAGUAAAGAAGACACAGUUG-UCGUUUUAUUAGAAUGAUUUACAUUUUAAUAAAGCAACAUUAAAGGUGUCUUCUUCCUUGU .....................((((((...))))))((((..((((((((((...((-(.((((((((((((((.....)))))))))))))).)))......)))))))))).)))). ( -33.10) >sy_ha.0 1710338 118 + 2685015 UUUUUUCCUCUCUAUUGUAAAAAUAUGUAAUAUAUACAAGUAAAGAAGACACUGACGCGCCUUCUAACAUAAAGUAUCACUUUAUAAU-AGAAAACCUCAAAGGUGUCUUCUUUCUUGU ...................................(((((.((((((((((((........(((((..((((((.....))))))..)-)))).........))))))))))))))))) ( -28.53) >consensus UCUUAUCCUCUCUAUUGUAAAAAUAUGUAAAAUAUACAAGUAAAGAAGACACAGUAG_GCCUUCUAAUACAAAGAUUAACAUUUUAAU_AAGAAACAUAAAAGGUGUCUUCUUCCUUGU ...................................(((((...(((((((((.............(((((((((.....)))))))))...............)))))))))..))))) (-16.96 = -17.86 + 0.90)

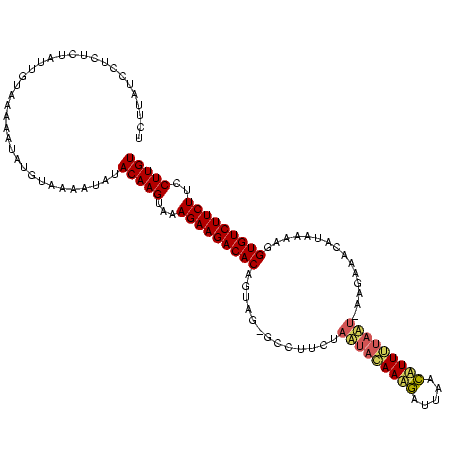
| Location | 1,569,641 – 1,569,757 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 74.37 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -20.19 |
| Energy contribution | -19.42 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.25 |
| Mean z-score | -4.19 |
| Structure conservation index | 0.62 |
| SVM decision value | 5.07 |
| SVM RNA-class probability | 0.999972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 1569641 116 + 2516575 ACAAGGAAGAAGACACCUUUUGUGCUCCGU--AUGAAAGAUUCACUUUUGUAUUGGAAGAC-CUAUUGUGUCUUCUUUACUUGUAUAAUUUACAUAUUUUUACAAUAGAGAGGAUAAGA ((((((((((((((((..(..((..(((((--(..((((.....))))..)).))))..))-..)..))))))))))).)))))..........(((((((........)))))))... ( -31.30) >sy_au.0 1522863 118 - 2809422 ACAAGGAAGAAGACACCUUUAAUGUUGCUUUAUUAAAAUGUAAAUCAUUCUAAUAAAACGA-CAACUGUGUCUUCUUUACUUGUAUAUGUUACAUAUAUUCACGAUAGAGAGGAUAAGA ((((((((((((((((......(((((.(((((((.((((.....)))).))))))).)))-))...))))))))))).)))))............(((((.(......).)))))... ( -32.00) >sy_ha.0 1710338 118 + 2685015 ACAAGAAAGAAGACACCUUUGAGGUUUUCU-AUUAUAAAGUGAUACUUUAUGUUAGAAGGCGCGUCAGUGUCUUCUUUACUUGUAUAUAUUACAUAUUUUUACAAUAGAGAGGAAAAAA ((((((((((((((((...(((((((((((-(.(((((((.....))))))).)))))))).).)))))))))))))).)))))................................... ( -35.10) >consensus ACAAGGAAGAAGACACCUUUAAUGUUCCCU_AUUAAAAAGUAAAACUUUAUAUUAGAAGGC_CAACUGUGUCUUCUUUACUUGUAUAUAUUACAUAUUUUUACAAUAGAGAGGAUAAGA ((((((((((((((((...............((((.((((.....)))).)))).............))))))))))).)))))..........(((((((.(....).)))))))... (-20.19 = -19.42 + -0.76) # Strand winner: reverse (0.98)

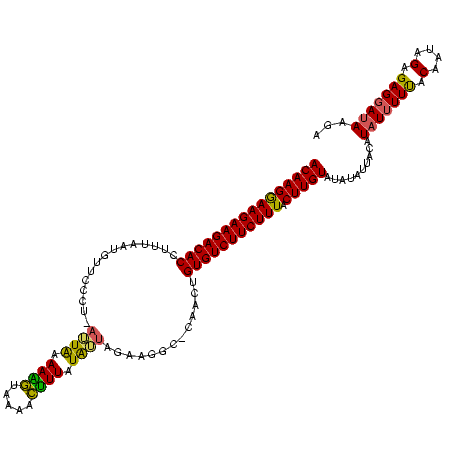
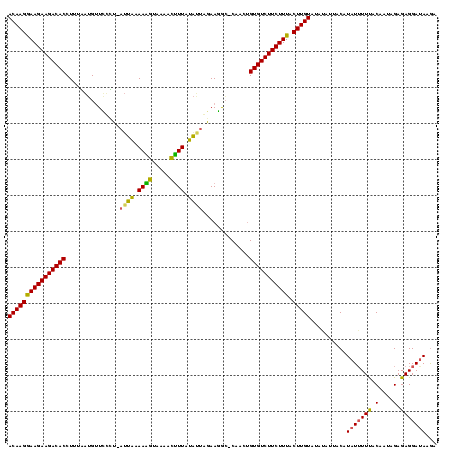
| Location | 1,569,641 – 1,569,757 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.37 |
| Mean single sequence MFE | -28.38 |
| Consensus MFE | -16.96 |
| Energy contribution | -17.86 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.96 |
| Structure conservation index | 0.60 |
| SVM decision value | 4.88 |
| SVM RNA-class probability | 0.999959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 1569641 116 + 2516575/0-119 UCUUAUCCUCUCUAUUGUAAAAAUAUGUAAAUUAUACAAGUAAAGAAGACACAAUAG-GUCUUCCAAUACAAAAGUGAAUCUUUCAU--ACGGAGCACAAAAGGUGUCUUCUUCCUUGU ...................................(((((..((((((((((.....-((.((((......((((.....))))...--..)))).)).....)))))))))).))))) ( -23.50) >sy_au.0 1522863 118 - 2809422/0-119 UCUUAUCCUCUCUAUCGUGAAUAUAUGUAACAUAUACAAGUAAAGAAGACACAGUUG-UCGUUUUAUUAGAAUGAUUUACAUUUUAAUAAAGCAACAUUAAAGGUGUCUUCUUCCUUGU .....................((((((...))))))((((..((((((((((...((-(.((((((((((((((.....)))))))))))))).)))......)))))))))).)))). ( -33.10) >sy_ha.0 1710338 118 + 2685015/0-119 UUUUUUCCUCUCUAUUGUAAAAAUAUGUAAUAUAUACAAGUAAAGAAGACACUGACGCGCCUUCUAACAUAAAGUAUCACUUUAUAAU-AGAAAACCUCAAAGGUGUCUUCUUUCUUGU ...................................(((((.((((((((((((........(((((..((((((.....))))))..)-)))).........))))))))))))))))) ( -28.53) >consensus UCUUAUCCUCUCUAUUGUAAAAAUAUGUAAAAUAUACAAGUAAAGAAGACACAGUAG_GCCUUCUAAUACAAAGAUUAACAUUUUAAU_AAGAAACAUAAAAGGUGUCUUCUUCCUUGU ...................................(((((...(((((((((.............(((((((((.....)))))))))...............)))))))))..))))) (-16.96 = -17.86 + 0.90)

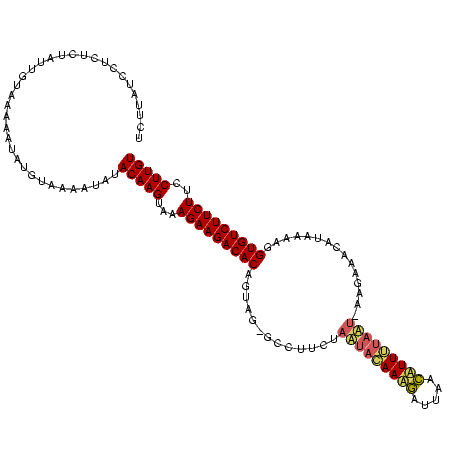

| Location | 1,569,641 – 1,569,757 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 74.37 |
| Mean single sequence MFE | -32.80 |
| Consensus MFE | -20.19 |
| Energy contribution | -19.42 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.25 |
| Mean z-score | -4.19 |
| Structure conservation index | 0.62 |
| SVM decision value | 5.07 |
| SVM RNA-class probability | 0.999972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 1569641 116 + 2516575/0-119 ACAAGGAAGAAGACACCUUUUGUGCUCCGU--AUGAAAGAUUCACUUUUGUAUUGGAAGAC-CUAUUGUGUCUUCUUUACUUGUAUAAUUUACAUAUUUUUACAAUAGAGAGGAUAAGA ((((((((((((((((..(..((..(((((--(..((((.....))))..)).))))..))-..)..))))))))))).)))))..........(((((((........)))))))... ( -31.30) >sy_au.0 1522863 118 - 2809422/0-119 ACAAGGAAGAAGACACCUUUAAUGUUGCUUUAUUAAAAUGUAAAUCAUUCUAAUAAAACGA-CAACUGUGUCUUCUUUACUUGUAUAUGUUACAUAUAUUCACGAUAGAGAGGAUAAGA ((((((((((((((((......(((((.(((((((.((((.....)))).))))))).)))-))...))))))))))).)))))............(((((.(......).)))))... ( -32.00) >sy_ha.0 1710338 118 + 2685015/0-119 ACAAGAAAGAAGACACCUUUGAGGUUUUCU-AUUAUAAAGUGAUACUUUAUGUUAGAAGGCGCGUCAGUGUCUUCUUUACUUGUAUAUAUUACAUAUUUUUACAAUAGAGAGGAAAAAA ((((((((((((((((...(((((((((((-(.(((((((.....))))))).)))))))).).)))))))))))))).)))))................................... ( -35.10) >consensus ACAAGGAAGAAGACACCUUUAAUGUUCCCU_AUUAAAAAGUAAAACUUUAUAUUAGAAGGC_CAACUGUGUCUUCUUUACUUGUAUAUAUUACAUAUUUUUACAAUAGAGAGGAUAAGA ((((((((((((((((...............((((.((((.....)))).)))).............))))))))))).)))))..........(((((((.(....).)))))))... (-20.19 = -19.42 + -0.76) # Strand winner: reverse (0.98)


Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:53:53 2006