
| Sequence ID | sy_au.0 |
|---|---|
| Location | 1,974,613 – 1,974,729 |
| Length | 116 |
| Max. P | 0.999216 |

| Location | 1,974,613 – 1,974,729 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 62.73 |
| Mean single sequence MFE | -29.93 |
| Consensus MFE | -15.55 |
| Energy contribution | -15.18 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.670442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1974613 116 + 2809422/0-120 UAUACCUCGUUCCGGGAAGGAACGUGUUCUAAAAGUUGAACUACUCCCGCAAAUAUUAA--AUUAUGGAGCGGAAGAUAGGAUUUACAUCUAUACCUCAUUCCAGGAAGGAAU--GUAUU ....(((((((((.....)))))).((((........))))..((.((((..(((....--..)))...)))).))..)))................((((((.....)))))--).... ( -27.70) >sy_ha.0 1574443 117 - 2685015/0-120 UAUACCUUGUUCCGGGAAGGAACAUAUUCUAAGAGUUGAAAUACUCCCGCAU-UAUUAAGAAUUAUGGAGCGGAAGAUAGGAUUUACACCUAUACCUUGUUCCGGGAAGGAAC--AUAUU .......((((((.....((((((........((((......))))((((.(-(((........)))).))))...(((((.......)))))....)))))).....)))))--).... ( -31.00) >sy_sa.0 1549552 112 - 2516575/0-120 UAUACCUCAUUCCGGGAAGGAAUGUGUUCUAAAAGUUGAACUACUCCCGCAA--AAUAA----UAUGGAGCGGAGGAUAGGACUUACACCUACACCUCGUUCCGGGAAGGAAC--GUGUU ....(((..((((((((.(......((((........))))..)))))....--.....----...((((((..(..((((.......))))..)..)))))))))))))...--..... ( -31.80) >sp_ag.0 2062975 107 - 2160267/0-120 AACGCAUGAGGGAGUCAAAGUCCCUUGCCUUACCGCUUGGCUAUACCC-----CAUGAA-------AAGGCGAGUGAUGGGAAUCGAACCCACGAAU-GUCAGAGCCACAAUCUGAUGUG ..(((..((((((.......))))))(((((....(.(((.......)-----)).)..-------)))))..))).((((.......))))...((-((((((.......)))))))). ( -29.20) >consensus UAUACCUCAUUCCGGGAAGGAACGUGUUCUAAAAGUUGAACUACUCCCGCAA_UAUUAA____UAUGGAGCGGAAGAUAGGAUUUACACCUACACCUCGUUCCAGGAAGGAAC__AUAUU ....(((((((((.....))))))..........................................(((((((...(((((.......)))))...)))))))))).............. (-15.55 = -15.18 + -0.38)

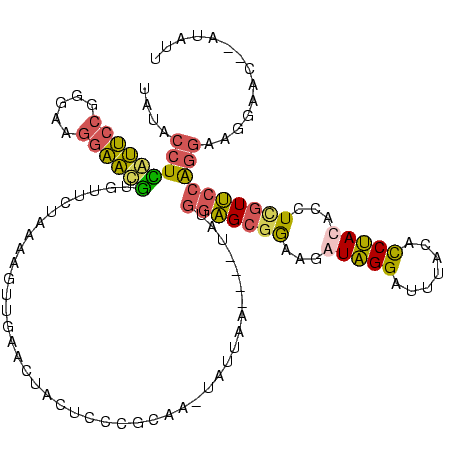
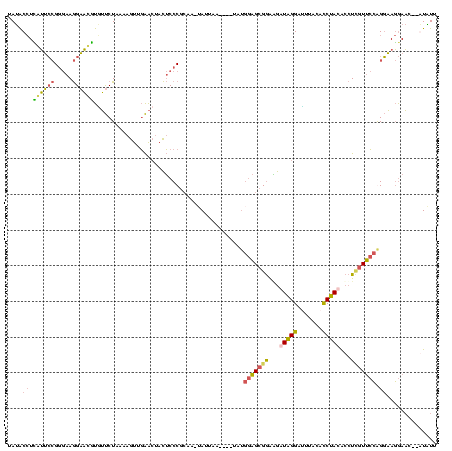
| Location | 1,974,613 – 1,974,729 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 62.73 |
| Mean single sequence MFE | -29.88 |
| Consensus MFE | -18.68 |
| Energy contribution | -17.69 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1974613 116 + 2809422/0-120 AAUAC--AUUCCUUCCUGGAAUGAGGUAUAGAUGUAAAUCCUAUCUUCCGCUCCAUAAU--UUAAUAUUUGCGGGAGUAGUUCAACUUUUAGAACACGUUCCUUCCCGGAACGAGGUAUA ....(--(((((.....))))))................(((..(((((((...(((..--....)))..)))))))..((((........)))).((((((.....))))))))).... ( -31.10) >sy_ha.0 1574443 117 - 2685015/0-120 AAUAU--GUUCCUUCCCGGAACAAGGUAUAGGUGUAAAUCCUAUCUUCCGCUCCAUAAUUCUUAAUA-AUGCGGGAGUAUUUCAACUCUUAGAAUAUGUUCCUUCCCGGAACAAGGUAUA ....(--(((((.....((((((.((.(((((.......)))))...))(((((...(((......)-))...)))))..................)))))).....))))))....... ( -28.70) >sy_sa.0 1549552 112 - 2516575/0-120 AACAC--GUUCCUUCCCGGAACGAGGUGUAGGUGUAAGUCCUAUCCUCCGCUCCAUA----UUAUU--UUGCGGGAGUAGUUCAACUUUUAGAACACAUUCCUUCCCGGAAUGAGGUAUA .....--(((((.....)))))((((.(((((.......))))))))).(((((...----.....--.....))))).((((........)))).((((((.....))))))....... ( -32.82) >sp_ag.0 2062975 107 - 2160267/0-120 CACAUCAGAUUGUGGCUCUGAC-AUUCGUGGGUUCGAUUCCCAUCACUCGCCUU-------UUCAUG-----GGGUAUAGCCAAGCGGUAAGGCAAGGGACUUUGACUCCCUCAUGCGUU ....(((((.......))))).-....(.(((.((((.((((.......(((((-------.....)-----))))...(((.........)))..))))..))))..))).)....... ( -26.90) >consensus AACAC__GUUCCUUCCCGGAACGAGGUAUAGGUGUAAAUCCUAUCUUCCGCUCCAUA____UUAAUA_UUGCGGGAGUAGUUCAACUUUUAGAACACGUUCCUUCCCGGAACGAGGUAUA .......(((((.....)))))((((((((((.......)))))(((((((...................))))))).......))))).......((((((.....))))))....... (-18.68 = -17.69 + -1.00) # Strand winner: reverse (0.91)

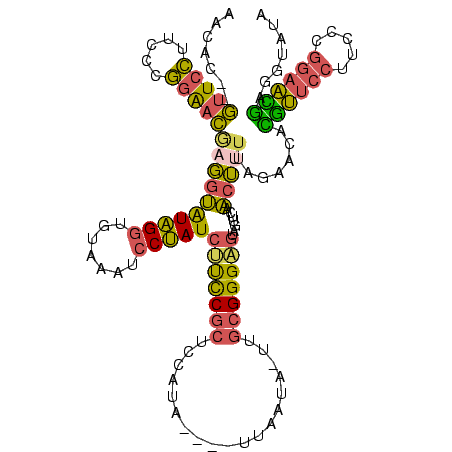
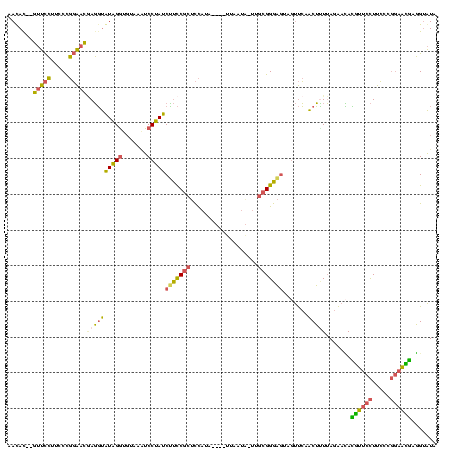
| Location | 1,974,613 – 1,974,723 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 63.36 |
| Mean single sequence MFE | -24.69 |
| Consensus MFE | -14.45 |
| Energy contribution | -13.33 |
| Covariance contribution | -1.12 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1974613 110 + 2809422/40-160 CUACUCCCGCAAAUAUUAA--AUUAUGGAGCGGAAGAUAGGAUUUACAUCUAUACCUCAUUCCAGGAAGGAAU--GUAUUCUAAGAGUUGAAAUACUCCCGCAU------UAUUAUUAAA ..................(--((.((((.((((....((((((..............((((((.....)))))--).)))))).((((......)))))))).)------))).)))... ( -19.56) >sy_ha.0 1574443 110 - 2685015/40-160 AUACUCCCGCAU-UAUUAAGAAUUAUGGAGCGGAAGAUAGGAUUUACACCUAUACCUUGUUCCGGGAAGGAAC--AUAUUCUAACAGUUGAAAUACUCCCGCAU------UAUUAA-GAA ........((..-.............(((((((...(((((.......)))))...)))))))((((.((((.--...))))....((......))))))))..------......-... ( -22.10) >sy_sa.0 1549552 112 - 2516575/40-160 CUACUCCCGCAA--AAUAA----UAUGGAGCGGAGGAUAGGACUUACACCUACACCUCGUUCCGGGAAGGAAC--GUGUUCUAAGAGUUGAACUACCCCCGCAUGGGAUAUAAUAAUUUA ....((((((..--.....----...((((((..(..((((.......))))..)..))))))(((..((...--..((((........))))..)))))))..))))............ ( -31.60) >sp_ag.0 2062975 101 - 2160267/40-160 CUAUACCC-----CAUGAA-------AAGGCGAGUGAUGGGAAUCGAACCCACGAAU-GUCAGAGCCACAAUCUGAUGUGUUAACCACUUCACCACACCCGCCA------UAUUAGAAAA ........-----......-------..((((.(((.(((((((((......)))((-((((((.......)))))))).........))).)))))).)))).------.......... ( -25.50) >consensus CUACUCCCGCAA_UAUUAA____UAUGGAGCGGAAGAUAGGAUUUACACCUACACCUCGUUCCAGGAAGGAAC__AUAUUCUAACAGUUGAAAUACUCCCGCAU______UAUUAAUAAA .............................((((.((.((((.......))))(((...(((((.....)))))..))).................)).)))).................. (-14.45 = -13.33 + -1.12)

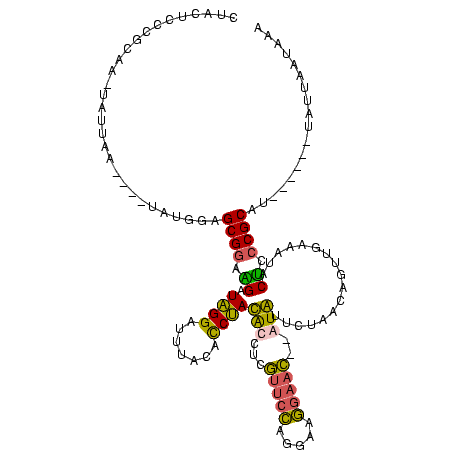
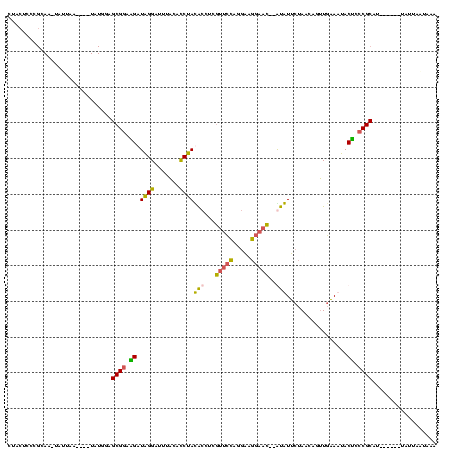
| Location | 1,974,613 – 1,974,723 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 63.36 |
| Mean single sequence MFE | -28.86 |
| Consensus MFE | -19.36 |
| Energy contribution | -17.55 |
| Covariance contribution | -1.81 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981825 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1974613 110 + 2809422/40-160 UUUAAUAAUA------AUGCGGGAGUAUUUCAACUCUUAGAAUAC--AUUCCUUCCUGGAAUGAGGUAUAGAUGUAAAUCCUAUCUUCCGCUCCAUAAU--UUAAUAUUUGCGGGAGUAG ..........------((((((((((......))))))......(--(((((.....))))))..)))).(((....)))(((.(((((((...(((..--....)))..)))))))))) ( -26.30) >sy_ha.0 1574443 110 - 2685015/40-160 UUC-UUAAUA------AUGCGGGAGUAUUUCAACUGUUAGAAUAU--GUUCCUUCCCGGAACAAGGUAUAGGUGUAAAUCCUAUCUUCCGCUCCAUAAUUCUUAAUA-AUGCGGGAGUAU ...-((((.(------((...(((((..................(--(((((.....)))))).((.(((((.......)))))...)))))))...))).))))..-((((....)))) ( -25.00) >sy_sa.0 1549552 112 - 2516575/40-160 UAAAUUAUUAUAUCCCAUGCGGGGGUAGUUCAACUCUUAGAACAC--GUUCCUUCCCGGAACGAGGUGUAGGUGUAAGUCCUAUCCUCCGCUCCAUA----UUAUU--UUGCGGGAGUAG ............((((..((((((((((..(.((.((((.(.(.(--(((((.....)))))).).).)))).))..)..)))))).))))..((..----.....--.)).)))).... ( -34.60) >sp_ag.0 2062975 101 - 2160267/40-160 UUUUCUAAUA------UGGCGGGUGUGGUGAAGUGGUUAACACAUCAGAUUGUGGCUCUGAC-AUUCGUGGGUUCGAUUCCCAUCACUCGCCUU-------UUCAUG-----GGGUAUAG .(..(.....------.((((((((((((((.(((.....))).)))(((((.(((((((..-...)).)))))))))).))).))))))))..-------.....)-----..)..... ( -29.52) >consensus UUUAUUAAUA______AUGCGGGAGUAGUUCAACUCUUAGAACAC__GUUCCUUCCCGGAACGAGGUAUAGGUGUAAAUCCUAUCUUCCGCUCCAUA____UUAAUA_UUGCGGGAGUAG ..................(((((((......................(((((.....))))).....(((((.......))))))))))))............................. (-19.36 = -17.55 + -1.81) # Strand winner: reverse (0.99)

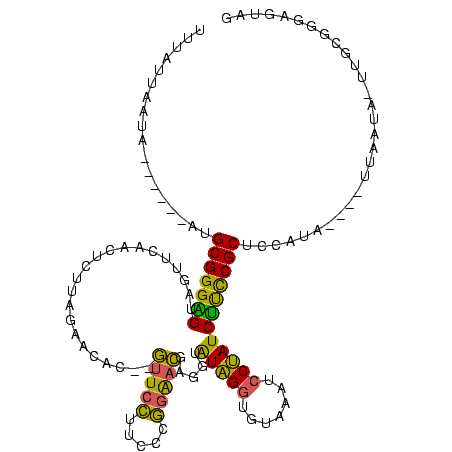
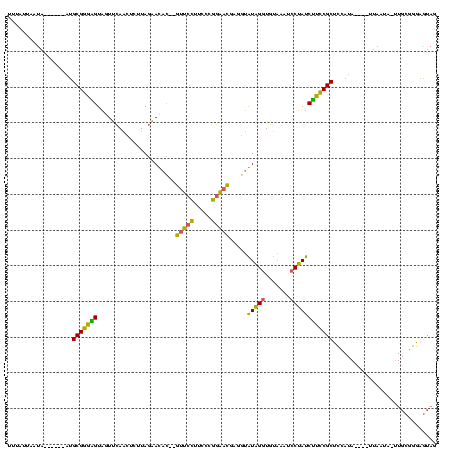
| Location | 1,974,613 – 1,974,725 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 64.51 |
| Mean single sequence MFE | -27.85 |
| Consensus MFE | -21.10 |
| Energy contribution | -18.29 |
| Covariance contribution | -2.81 |
| Combinations/Pair | 1.62 |
| Mean z-score | -0.64 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1974613 112 + 2809422/80-200 GAACGAGGUAUAGGUGCAAAUCCUAUCUUCCGCUCCAUAAUUUAAUAAUA------AUGCGGGAGUAUUUCAACUCUUAGAAUAC--AUUCCUUCCUGGAAUGAGGUAUAGAUGUAAAUC ....((((((((((.......)))).(((((((.................------..))))))))))))).(((((.......(--(((((.....))))))......))).))..... ( -24.53) >sy_ha.0 1574443 111 - 2685015/80-200 GAAUGAGGUAUAGGUGCAAGUCCUAUCUUCCGCUCCAUAAUUC-UUAAUA------AUGCGGGAGUAUUUCAACUGUUAGAAUAU--GUUCCUUCCCGGAACAAGGUAUAGGUGUAAAUC ...(((((((((((.......)))).(((((((..........-......------..)))))))))))))).((((.......(--(((((.....))))))....))))......... ( -25.07) >sy_sa.0 1549552 118 - 2516575/80-200 GAAUGAGGUAUAGGUGUAAGUCCUAUCUUCCGCUCCAUAUUAAAUUAUUAUAUCCCAUGCGGGGGUAGUUCAACUCUUAGAACAC--GUUCCUUCCCGGAACGAGGUGUAGGUGUAAGUC ....((((.(((((.......))))))))).((.(((((((.........((((((.....))))))((((........)))).(--(((((.....)))))).))))).)).))..... ( -31.70) >sp_ag.0 2062975 109 - 2160267/80-200 ACCUUCAGUGUGGGUUCGAUUCCUACUGCCCGUGUU----UUUUCUAAUA------UGGCGGGUGUGGUGAAGUGGUUAACACAUCAGAUUGUGGCUCUGAC-AUUCGUGGGUUCGAUUC ((((.......))))((((..(((((.(((.(((((----......))))------))))((((((......(((.....)))..((((.......))))))-))))))))).))))... ( -30.10) >consensus GAAUGAGGUAUAGGUGCAAGUCCUAUCUUCCGCUCCAUAAUUUAUUAAUA______AUGCGGGAGUAGUUCAACUCUUAGAACAC__GUUCCUUCCCGGAACGAGGUAUAGGUGUAAAUC ....((((((((((.......)))))(((((((.........................))))))).)))))..........((((..(((((.....)))))...))))........... (-21.10 = -18.29 + -2.81) # Strand winner: reverse (1.00)

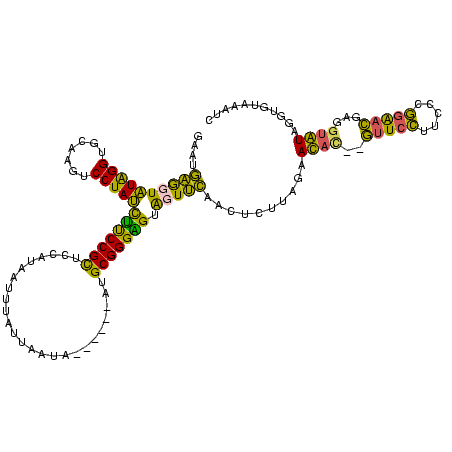
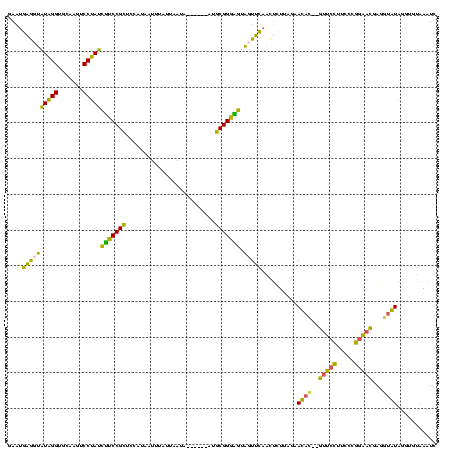
| Location | 1,974,613 – 1,974,727 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.04 |
| Mean single sequence MFE | -24.85 |
| Consensus MFE | -22.04 |
| Energy contribution | -19.35 |
| Covariance contribution | -2.69 |
| Combinations/Pair | 1.48 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968171 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1974613 114 + 2809422/120-240 CUAAGAGUUGAAAUACUCCCGCAU------UAUUAUUAAAUUAUGGAGCGGAAGAUAGGAUUUGCACCUAUACCUCGUUCCGGGAAGGAACGUGUUCUAAAAGUUGAACUACUCCCGCAU ....((((......))))..((..------..............(((((((...(((((.......)))))...)))))))((((.(......((((........))))..))))))).. ( -29.70) >sy_ha.0 1574443 112 - 2685015/120-240 CUAACAGUUGAAAUACUCCCGCAU------UAUUAA-GAAUUAUGGAGCGGAAGAUAGGACUUGCACCUAUACCUCAUUCCGGGAAGGAAUGUGUUCUAAAA-UUGAACUACUCCCGCAU ................(((...((------(.....-.)))...)))((((.(((((((.......)))))....((((((.....)))))).((((.....-..))))..)).)))).. ( -22.70) >sy_sa.0 1549552 120 - 2516575/120-240 CUAAGAGUUGAACUACCCCCGCAUGGGAUAUAAUAAUUUAAUAUGGAGCGGAAGAUAGGACUUACACCUAUACCUCAUUCCGGGAAGGAACGUGUUCUAAAAGUUGAACUACUCCCGCAU .....((((.((((.(((......)))................((((((((...(((((.......))))).))...((((.....))))...))))))..)))).)))).......... ( -26.20) >sp_ag.0 2062975 109 - 2160267/120-240 UUAACCACUUCACCACACCCGCCA------UAUUAGAAAA----AACACGGGCAGUAGGAAUCGAACCCACACUGAAGGUUUUGGAGACCUUAGUUCUACCU-UUAAACUAUGCCCGUUU ........................------..........----...(((((((((.((........))))(((((.((((.....))))))))).......-........))))))).. ( -20.80) >consensus CUAACAGUUGAAAUACUCCCGCAU______UAUUAAUAAAUUAUGGAGCGGAAGAUAGGACUUGCACCUAUACCUCAUUCCGGGAAGGAACGUGUUCUAAAA_UUGAACUACUCCCGCAU ...............................................((((.(((((((.......)))))....((((((.....)))))).((((........))))..)).)))).. (-22.04 = -19.35 + -2.69)

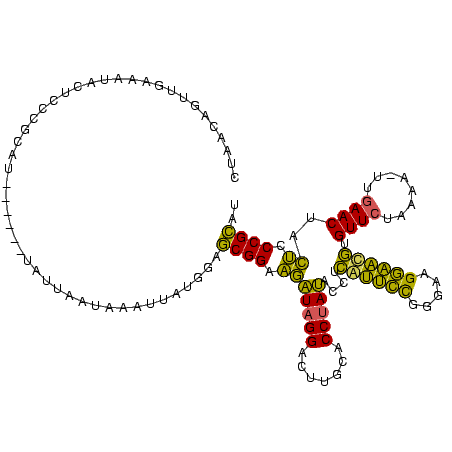
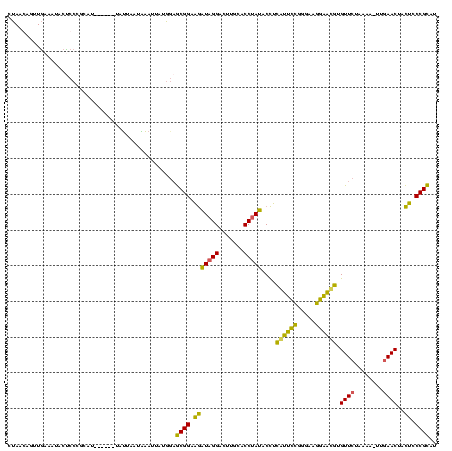
| Location | 1,974,613 – 1,974,727 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.04 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -29.15 |
| Energy contribution | -25.02 |
| Covariance contribution | -4.12 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.44 |
| SVM RNA-class probability | 0.999216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1974613 114 + 2809422/120-240 AUGCGGGAGUAGUUCAACUUUUAGAACACGUUCCUUCCCGGAACGAGGUAUAGGUGCAAAUCCUAUCUUCCGCUCCAUAAUUUAAUAAUA------AUGCGGGAGUAUUUCAACUCUUAG ....((((((.((((........))))..(((((.....)))))((((((((((.......)))).(((((((.................------..))))))))))))).)))))).. ( -31.31) >sy_ha.0 1574443 112 - 2685015/120-240 AUGCGGGAGUAGUUCAA-UUUUAGAACACAUUCCUUCCCGGAAUGAGGUAUAGGUGCAAGUCCUAUCUUCCGCUCCAUAAUUC-UUAAUA------AUGCGGGAGUAUUUCAACUGUUAG ..(((((((..((((..-.....)))).((((((.....))))))....(((((.......))))))))))))....((((..-.....(------((((....)))))......)))). ( -28.82) >sy_sa.0 1549552 120 - 2516575/120-240 AUGCGGGAGUAGUUCAACUUUUAGAACACGUUCCUUCCCGGAAUGAGGUAUAGGUGUAAGUCCUAUCUUCCGCUCCAUAUUAAAUUAUUAUAUCCCAUGCGGGGGUAGUUCAACUCUUAG ..(((((((..((((........)))).((((((.....))))))....(((((.......)))))))))))).................((((((.....))))))............. ( -31.40) >sp_ag.0 2062975 109 - 2160267/120-240 AAACGGGCAUAGUUUAA-AGGUAGAACUAAGGUCUCCAAAACCUUCAGUGUGGGUUCGAUUCCUACUGCCCGUGUU----UUUUCUAAUA------UGGCGGGUGUGGUGAAGUGGUUAA ((((.......))))..-......(((((...((.(((..((((.(((((.(((......)))))))).(((((((----......))))------))).)))).))).))..))))).. ( -25.80) >consensus AUGCGGGAGUAGUUCAA_UUUUAGAACACAUUCCUUCCCGGAAUGAGGUAUAGGUGCAAGUCCUAUCUUCCGCUCCAUAAUUUAUUAAUA______AUGCGGGAGUAGUUCAACUCUUAG ..(((((((..((((........)))).((((((.....))))))....(((((.......))))))))))))...........................((((((......)))))).. (-29.15 = -25.02 + -4.12) # Strand winner: reverse (1.00)

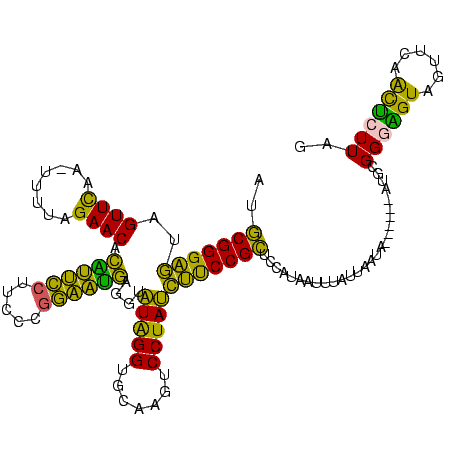
| Location | 1,974,613 – 1,974,729 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.61 |
| Mean single sequence MFE | -34.62 |
| Consensus MFE | -29.95 |
| Energy contribution | -27.20 |
| Covariance contribution | -2.75 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.02 |
| SVM RNA-class probability | 0.998173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1974613 116 + 2809422/160-280 AUCACCGGUUCAAA---UCCGGUUGCCGCCUCCAGGU-UUAUGCGGGAGUAGUUCAACUUUUAGAACACGUUCCUUCCCGGAACGAGGUAUAGGUGCAAAUCCUAUCUUCCGCUCCAUAA ...(((((......---.)))))(((.(((....)))-....)))(((((.((((........))))..(((((.....)))))((((.(((((.......))))))))).))))).... ( -35.50) >sy_ha.0 1574443 116 - 2685015/160-280 AUCACCGGUUCAAA---UCCGGUUGCCGCCUCCAUGCACGAUGCGGGAGUAGUUCAA-UUUUAGAACACAUUCCUUCCCGGAAUGAGGUAUAGGUGCAAGUCCUAUCUUCCGCUCCAUAA ...(((((......---.)))))...(((.((.......)).)))(((((.((((..-.....)))).((((((.....)))))).((.(((((.......)))))...))))))).... ( -34.00) >sy_sa.0 1549552 116 - 2516575/160-280 AUCACCGGUUCAAA---UCCGGUUGCCGCCUUCA-ACAAUAUGCGGGAGUAGUUCAACUUUUAGAACACGUUCCUUCCCGGAAUGAGGUAUAGGUGUAAGUCCUAUCUUCCGCUCCAUAU ...(((((......---.)))))...(((.....-.......)))(((((.((((........)))).((((((.....)))))).((.(((((.......)))))...))))))).... ( -31.70) >sp_ag.0 2062975 115 - 2160267/160-280 CCUUCGGGUUCGGGGGUUCGAAUCCCUCUCUCUCCAUAGUAAACGGGCAUAGUUUAA-AGGUAGAACUAAGGUCUCCAAAACCUUCAGUGUGGGUUCGAUUCCUACUGCCCGUGUU---- ....(((((...(((..((((((((.....(((((....(((((.......))))).-.)).)))((((((((.......))))).)))..))))))))..)))...)))))....---- ( -37.30) >consensus AUCACCGGUUCAAA___UCCGGUUGCCGCCUCCA_ACAAUAUGCGGGAGUAGUUCAA_UUUUAGAACACAUUCCUUCCCGGAAUGAGGUAUAGGUGCAAGUCCUAUCUUCCGCUCCAUAA ...(((((..........)))))...................(((((((..((((........)))).((((((.....))))))....(((((.......))))))))))))....... (-29.95 = -27.20 + -2.75) # Strand winner: reverse (0.99)

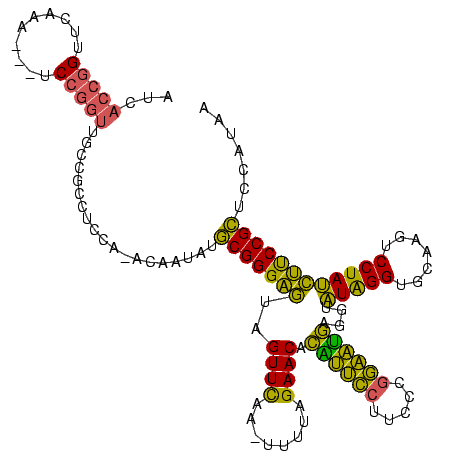
| Location | 1,974,613 – 1,974,721 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.37 |
| Mean single sequence MFE | -36.85 |
| Consensus MFE | -23.29 |
| Energy contribution | -22.10 |
| Covariance contribution | -1.19 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1974613 108 + 2809422/238-358 AUAA-ACCUGGAGGCGGCAACCGGA---UUUGAACCGGUGAUAAAGGUUUUGCAGACCUCUGCCUU--ACCACUUGGCUAUGCCGCCAAUAA------CUGGGCUAGCUGGAUUCGAACC ....-.((..(.((((((((((((.---......)))))......(((...((((....))))...--))).........))))))).....------)..))................. ( -34.70) >sy_ha.0 1574443 109 - 2685015/238-358 AUCGUGCAUGGAGGCGGCAACCGGA---UUUGAACCGGUGAUAAAGGUUUUGCAGACCUCUGCCUU--ACCACUUGGCUAUGCCGCCAAUAA------CUGGGCUAGCUGGAUUCGAACC .(((........((((((((((((.---......)))))......(((...((((....))))...--))).........))))))).....------(..(.....)..)...)))... ( -35.30) >sy_sa.0 1549552 112 - 2516575/238-358 AUAUUGU-UGAAGGCGGCAACCGGA---UUUGAACCGGUGAUAAAGGUUUUGCAGACCUCUGCCUU--ACCACUUGGCUAUGCCGCCAAUAAAA--AACUGGGCUAGCUGGAUUCGAACC ......(-((((((((((((((((.---......)))))......(((...((((....))))...--))).........))))))).......--..(..(.....)..).)))))... ( -36.30) >sp_ag.0 2062975 120 - 2160267/238-358 UUACUAUGGAGAGAGAGGGAUUCGAACCCCCGAACCCGAAGGAGCGGAUUUACAGUCCGCCGCGUUUAGCCUCUUCGCUAUCUCUCCUAAGGUAUAAAUGGCGCGAGACGGAAUCGAACC .((((..(((((((..(((.((((......)))))))(((((.((((((.....)))))).((.....)).)))))....)))))))...)))).....((..(((.......)))..)) ( -41.10) >consensus AUAAUAC_UGGAGGCGGCAACCGGA___UUUGAACCGGUGAUAAAGGUUUUGCAGACCUCUGCCUU__ACCACUUGGCUAUGCCGCCAAUAA______CUGGGCUAGCUGGAUUCGAACC ............((((((((((((..........))))).....(((((.....)))))..(((...........)))..)))))))................................. (-23.29 = -22.10 + -1.19)

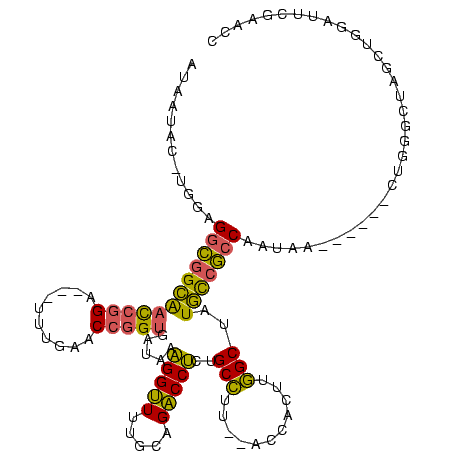
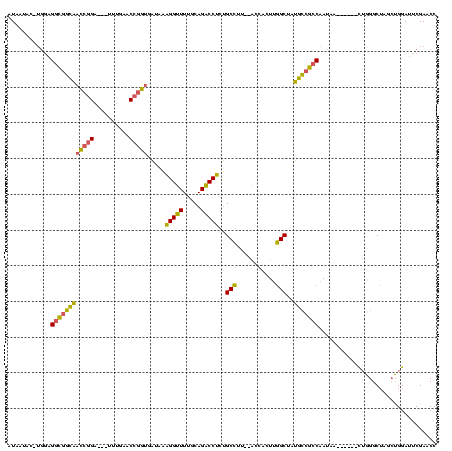
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:53:49 2006