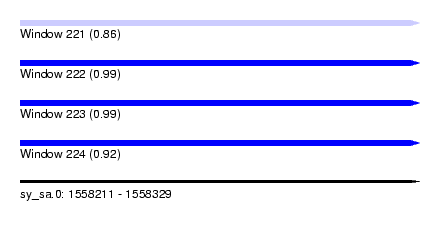
| Sequence ID | sy_sa.0 |
|---|---|
| Location | 1,558,211 – 1,558,329 |
| Length | 118 |
| Max. P | 0.986937 |

| Location | 1,558,211 – 1,558,329 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.55 |
| Mean single sequence MFE | -23.80 |
| Consensus MFE | -16.51 |
| Energy contribution | -16.40 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 1558211 118 + 2516575/0-120 GACCUCCUUUUUUGGCAAAUAGAAAAGAGCGGGUAUUAUGCCCACUCUUUCUGCCUGAGUCUAAU--UUUUUAAGCACUAUUAUUAUACCAUAUGUUCAUUUUCUAUACAAAUAACUAUA ..........((((....((((((((((((((((.....))))........(((.((((......--..)))).))).................)))).)))))))).))))........ ( -20.70) >sy_ha.0 1699301 117 + 2685015/0-120 GGCCUCCUUUUUCGGCAAAUAGAAAAGAGCGGGUAU-AUGCCCACUCUUUCUGCCUGAGUCUAAU--UUUUUACGCAACUUAAGUAUAUCACACUUUAAUUCCUUAAACAAAUGUCUAUA .(((.........)))..(((((((((((.((((..-..)))).)))))).(((.((((......--..)))).))).....................................))))). ( -22.60) >sy_au.0 1511527 117 - 2809422/0-120 GGCCUCCCUUUUUGGCAAAUAGAAAAGAGCGGGAAU--CUCCCACUCUU-CUGCCUGAGUUCACUAAUUUUUAAGCAACUUAAUUAUAGCAUAAGUUUAUGCUUGAAACAAAUGACUUCA .(((.........)))...((((..((((.(((...--..))).)))))-)))...(((.(((.....((((((((((((((.........)))))...)))))))))....))).))). ( -28.10) >consensus GGCCUCCUUUUUUGGCAAAUAGAAAAGAGCGGGUAU_AUGCCCACUCUUUCUGCCUGAGUCUAAU__UUUUUAAGCAACUUAAUUAUACCAUAAGUUAAUUCUUUAAACAAAUGACUAUA .............((((......((((((.((((.....)))).)))))).))))................................................................. (-16.51 = -16.40 + -0.11)

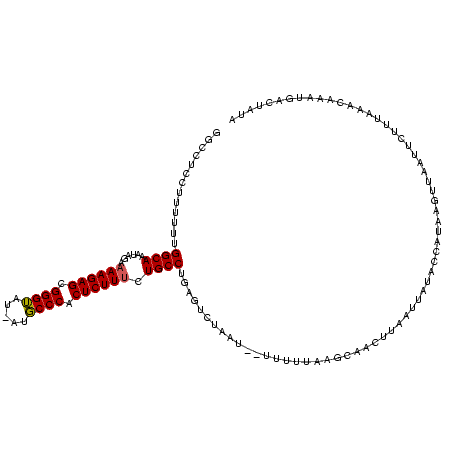
| Location | 1,558,211 – 1,558,329 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.55 |
| Mean single sequence MFE | -30.45 |
| Consensus MFE | -20.93 |
| Energy contribution | -21.27 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 1558211 118 + 2516575/0-120 UAUAGUUAUUUGUAUAGAAAAUGAACAUAUGGUAUAAUAAUAGUGCUUAAAAA--AUUAGACUCAGGCAGAAAGAGUGGGCAUAAUACCCGCUCUUUUCUAUUUGCCAAAAAAGGAGGUC (((..((((((.......))))))..)))(((((....((((((((((.....--.........)))))(((((((((((.......)))))))))))))))))))))............ ( -25.54) >sy_ha.0 1699301 117 + 2685015/0-120 UAUAGACAUUUGUUUAAGGAAUUAAAGUGUGAUAUACUUAAGUUGCGUAAAAA--AUUAGACUCAGGCAGAAAGAGUGGGCAU-AUACCCGCUCUUUUCUAUUUGCCGAAAAAGGAGGCC ..(((((....))))).(.((((.((((((...)))))).)))).).......--......(((.(((((((((((((((...-...))))))))))).....)))).......)))... ( -30.60) >sy_au.0 1511527 117 - 2809422/0-120 UGAAGUCAUUUGUUUCAAGCAUAAACUUAUGCUAUAAUUAAGUUGCUUAAAAAUUAGUGAACUCAGGCAG-AAGAGUGGGAG--AUUCCCGCUCUUUUCUAUUUGCCAAAAAGGGAGGCC (((..(((((..(((.(((((...(((((.........)))))))))).)))...)))))..)))(((((-(((((((((..--...)))))))))))....((.((.....)).))))) ( -35.20) >consensus UAUAGUCAUUUGUUUAAAGAAUAAACAUAUGAUAUAAUUAAGUUGCUUAAAAA__AUUAGACUCAGGCAGAAAGAGUGGGCAU_AUACCCGCUCUUUUCUAUUUGCCAAAAAAGGAGGCC .............................................................(((.(((((((((((((((.......))))))))).....)))))).......)))... (-20.93 = -21.27 + 0.33) # Strand winner: reverse (1.00)


| Location | 1,558,211 – 1,558,329 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.12 |
| Mean single sequence MFE | -21.67 |
| Consensus MFE | -13.21 |
| Energy contribution | -12.77 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
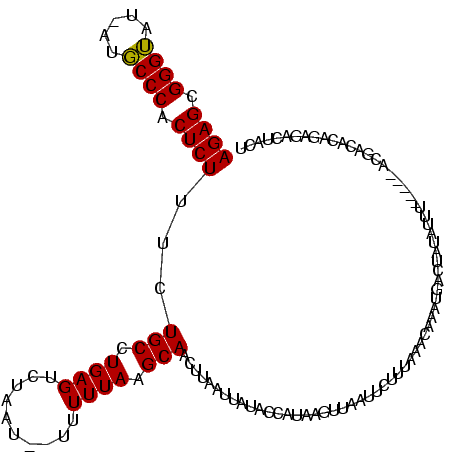
>sy_sa.0 1558211 118 + 2516575/25-145 AGAGCGGGUAUUAUGCCCACUCUUUCUGCCUGAGUCUAAU--UUUUUAAGCACUAUUAUUAUACCAUAUGUUCAUUUUCUAUACAAAUAACUAUAUUUGUUAAACGGCACAAUGACAACU ((((.((((.....)))).))))...((((...(..(((.--...)))..)...............................(((((((....))))))).....))))........... ( -20.20) >sy_ha.0 1699301 113 + 2685015/25-145 AGAGCGGGUAU-AUGCCCACUCUUUCUGCCUGAGUCUAAU--UUUUUACGCAACUUAAGUAUAUCACACUUUAAUUCCUUAAACAAAUGUCUAUAUCU----UGCGUCACAGAGACUACU ((((.((((..-..)))).)))).........(((((...--.....((((((..((((.((((.....(((((....)))))...)))))).))..)----))))).....)))))... ( -21.62) >sy_au.0 1511527 111 - 2809422/25-145 AGAGCGGGAAU--CUCCCACUCUU-CUGCCUGAGUUCACUAAUUUUUAAGCAACUUAAUUAUAGCAUAAGUUUAUGCUUGAAACAAAUGACUUCACU------AUUAAUCAGAGAUUCUU .(((.(((...--..))).)))((-(((..((((.(((.....((((((((((((((.........)))))...)))))))))....))).))))..------......)))))...... ( -23.20) >consensus AGAGCGGGUAU_AUGCCCACUCUUUCUGCCUGAGUCUAAU__UUUUUAAGCAACUUAAUUAUACCAUAAGUUAAUUCUUUAAACAAAUGACUAUAUUU_____ACGACACAGAGACUACU ((((.((((.....)))).))))...(((.((((..........)))).))).................................................................... (-13.21 = -12.77 + -0.44)

| Location | 1,558,211 – 1,558,329 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.12 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -13.19 |
| Energy contribution | -13.52 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
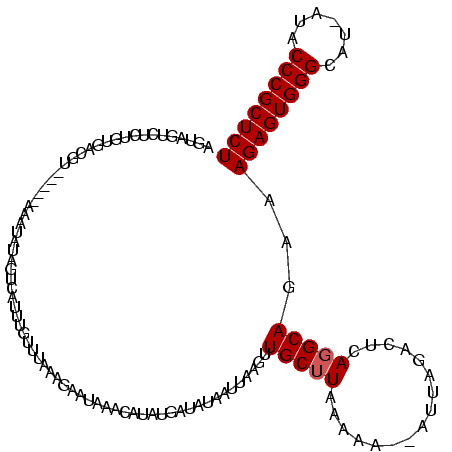
>sy_sa.0 1558211 118 + 2516575/25-145 AGUUGUCAUUGUGCCGUUUAACAAAUAUAGUUAUUUGUAUAGAAAAUGAACAUAUGGUAUAAUAAUAGUGCUUAAAAA--AUUAGACUCAGGCAGAAAGAGUGGGCAUAAUACCCGCUCU ..(((((((((((((((((((((((((....)))))))........)))))....))))))))...(((.((......--...)))))..)))))..((((((((.......)))))))) ( -27.60) >sy_ha.0 1699301 113 + 2685015/25-145 AGUAGUCUCUGUGACGCA----AGAUAUAGACAUUUGUUUAAGGAAUUAAAGUGUGAUAUACUUAAGUUGCGUAAAAA--AUUAGACUCAGGCAGAAAGAGUGGGCAU-AUACCCGCUCU .(.((((((((((.(...----.).))))))......((((.(.((((.((((((...)))))).)))).).))))..--....)))))........((((((((...-...)))))))) ( -29.20) >sy_au.0 1511527 111 - 2809422/25-145 AAGAAUCUCUGAUUAAU------AGUGAAGUCAUUUGUUUCAAGCAUAAACUUAUGCUAUAAUUAAGUUGCUUAAAAAUUAGUGAACUCAGGCAG-AAGAGUGGGAG--AUUCCCGCUCU .......((((......------..(((..(((((..(((.(((((...(((((.........)))))))))).)))...)))))..)))..)))-)((((((((..--...)))))))) ( -28.60) >consensus AGUAGUCUCUGUGACGU_____AAAUAUAGUCAUUUGUUUAAAGAAUAAACAUAUGAUAUAAUUAAGUUGCUUAAAAA__AUUAGACUCAGGCAGAAAGAGUGGGCAU_AUACCCGCUCU ....................................................................(((((................)))))...((((((((.......)))))))) (-13.19 = -13.52 + 0.33) # Strand winner: reverse (0.76)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:53:41 2006