
| Sequence ID | sp_mu.0 |
|---|---|
| Location | 1,839,762 – 1,839,882 |
| Length | 120 |
| Max. P | 0.991808 |

| Location | 1,839,762 – 1,839,879 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.56 |
| Mean single sequence MFE | -34.40 |
| Consensus MFE | -29.20 |
| Energy contribution | -29.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.962383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
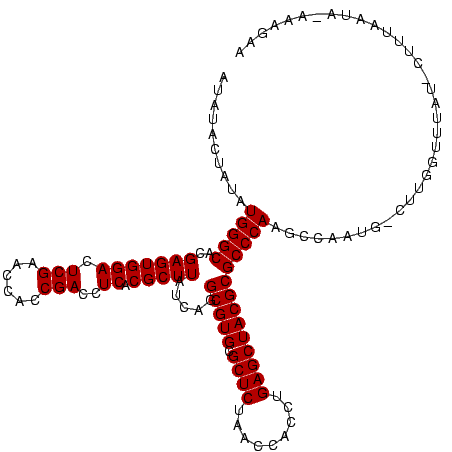
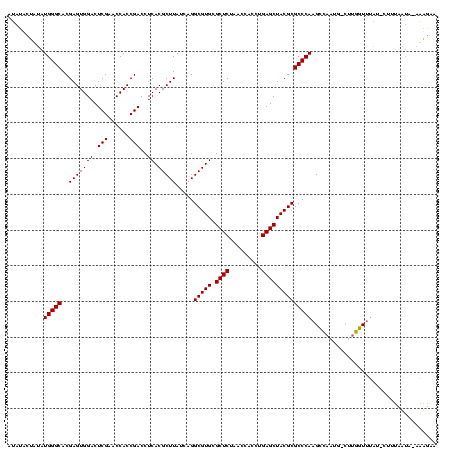
>sp_mu.0 1839762 117 - 2030921/0-120 AUAUACUAUAUGGGCACGAGUGGACUCGAACCACCGACCUCACGCUUAUCAGGCGUGCGCUCUAACCAGCUGAGCUACGCGCCCAAGCCUAUG-CUUGGUUUAU-CUUUUAUA-AAAGAA .((.(((((((((((..(((((((.(((......)))..)).)))))....((((((.((((.........))))..))))))...)))))))-..)))).))(-((((....-))))). ( -34.10) >sp_ag.0 1904703 119 - 2160267/0-120 AUAUUCUUGAUGGGCACGAGUGGACUCGAACCACCGACCUCACGCUUAUCAGGCGUGCGCUCUAACCACCUGAGCUACGCGCCCAAGCUAUUG-CUUGGUUUUUACUUUCUUAUAAAGUA ......(((((((((....((((..(((......)))..)))))))))))))(((((.((((.........)))))))))..((((((....)-)))))....((((((.....)))))) ( -37.50) >sp_pn.0 1851494 117 + 2038615/0-120 CUAAACACUAUGGGCACGAGUGGACUCGAACCACCGACCUCACGCUUAUCAGGCGUGCGCUCUAACCACCUGAGCUACGCGCCCAAGUCAAAAACUUGGUACAA---AGAACAAAGUUCA ......(((.(((((..(((((((.(((......)))..)).))))).....(((((.((((.........)))))))))))))))))....(((((.((....---...)).))))).. ( -31.60) >consensus AUAUACUAUAUGGGCACGAGUGGACUCGAACCACCGACCUCACGCUUAUCAGGCGUGCGCUCUAACCACCUGAGCUACGCGCCCAAGCCAAUG_CUUGGUUUAU_CUUUAAUA_AAAGAA ..........(((((..(((((((.(((......)))..)).))))).....(((((.((((.........))))))))))))))................................... (-29.20 = -29.20 + -0.00)

| Location | 1,839,762 – 1,839,879 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.56 |
| Mean single sequence MFE | -39.53 |
| Consensus MFE | -32.39 |
| Energy contribution | -32.50 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811778 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
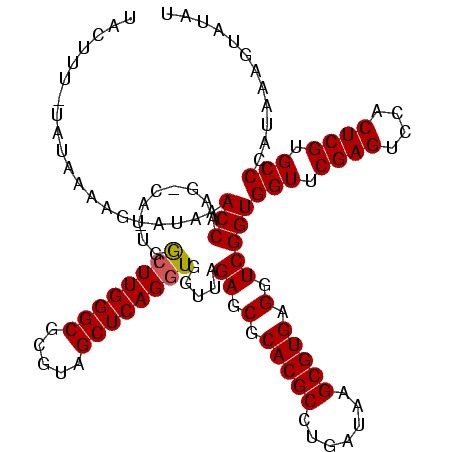
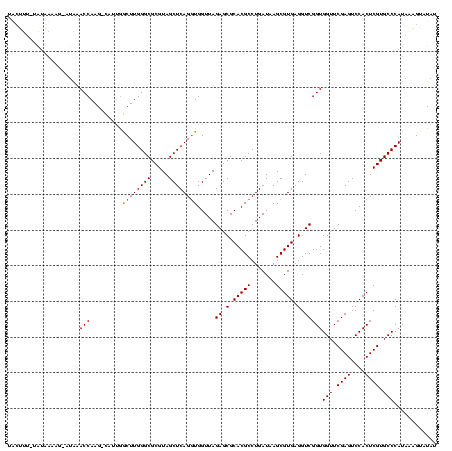
>sp_mu.0 1839762 117 - 2030921/0-120 UUCUUU-UAUAAAAG-AUAAACCAAG-CAUAGGCUUGGGCGCGUAGCUCAGCUGGUUAGAGCGCACGCCUGAUAAGCGUGAGGUCGGUGGUUCGAGUCCACUCGUGCCCAUAUAGUAUAU .(((((-....))))-)......(((-(....))))(((((((...(((((((.(((((.((....))))))).))).))))...(((((.......))))))))))))........... ( -38.60) >sp_ag.0 1904703 119 - 2160267/0-120 UACUUUAUAAGAAAGUAAAAACCAAG-CAAUAGCUUGGGCGCGUAGCUCAGGUGGUUAGAGCGCACGCCUGAUAAGCGUGAGGUCGGUGGUUCGAGUCCACUCGUGCCCAUCAAGAAUAU ((((((.....))))))....(((((-(....)))))).(((((...(((((((((......)).)))))))...))))).....(((((..((((....))))...)))))........ ( -41.30) >sp_pn.0 1851494 117 + 2038615/0-120 UGAACUUUGUUCU---UUGUACCAAGUUUUUGACUUGGGCGCGUAGCUCAGGUGGUUAGAGCGCACGCCUGAUAAGCGUGAGGUCGGUGGUUCGAGUCCACUCGUGCCCAUAGUGUUUAG .((((((.((...---....)).))))))...(((((((((((..(((((((((((......)).)))))))...))(((.(.((((....)))).).))).)))))))).)))...... ( -38.70) >consensus UACUUU_UAUAAAAG_AUAAACCAAG_CAUUGGCUUGGGCGCGUAGCUCAGGUGGUUAGAGCGCACGCCUGAUAAGCGUGAGGUCGGUGGUUCGAGUCCACUCGUGCCCAUAAAGUAUAU ....................(((.........((((((((.....)))))))).....((.(.(((((.......))))).).)))))(((.((((....)))).)))............ (-32.39 = -32.50 + 0.11) # Strand winner: reverse (0.99)

| Location | 1,839,762 – 1,839,879 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.20 |
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -31.12 |
| Energy contribution | -30.13 |
| Covariance contribution | -0.99 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.800101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 1839762 117 - 2030921/40-160 CACGCUUAUCAGGCGUGCGCUCUAACCAGCUGAGCUACGCGCCCAAGCCUAUG-CUUGGUUUAU-CUUUUAUA-AAAGAAAAGCGGGUGACGAGAAUCGAACUCGCGACAACAGCUUGGA ((((((.....))))))........(((((((...(((.(((((((((....)-)))))....(-((((....-)))))...))).))).((((.......))))......)))).))). ( -37.50) >sp_ag.0 1904703 119 - 2160267/40-160 CACGCUUAUCAGGCGUGCGCUCUAACCACCUGAGCUACGCGCCCAAGCUAUUG-CUUGGUUUUUACUUUCUUAUAAAGUAAAGCGGGUGACGAGAAUCGAACUCGCGACAACAGCUUGGA ((((((.....)))))).((((.........)))).......(((((((.(((-(......((((((((.....))))))))((((((..((.....)).))))))).))).))))))). ( -40.40) >sp_pn.0 1851494 117 + 2038615/40-160 CACGCUUAUCAGGCGUGCGCUCUAACCACCUGAGCUACGCGCCCAAGUCAAAAACUUGGUACAA---AGAACAAAGUUCAAAGCGGGUGACGAGAAUCGAACUCGCGACAACAGCUUGGA ((.(((.....((((((.((((.........))))..))))))...(((...(((((.((....---...)).)))))....((((((..((.....)).)))))))))...))).)).. ( -34.30) >consensus CACGCUUAUCAGGCGUGCGCUCUAACCACCUGAGCUACGCGCCCAAGCCAAUG_CUUGGUUUAU_CUUUAAUA_AAAGAAAAGCGGGUGACGAGAAUCGAACUCGCGACAACAGCUUGGA ((((((.....)))))).((((.........)))).......((((((....................((((...))))...((((((..((.....)).)))))).......)))))). (-31.12 = -30.13 + -0.99)

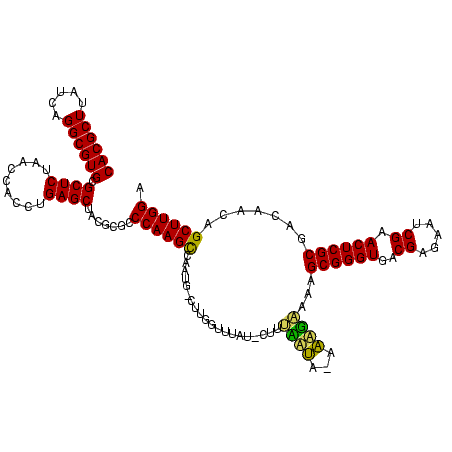
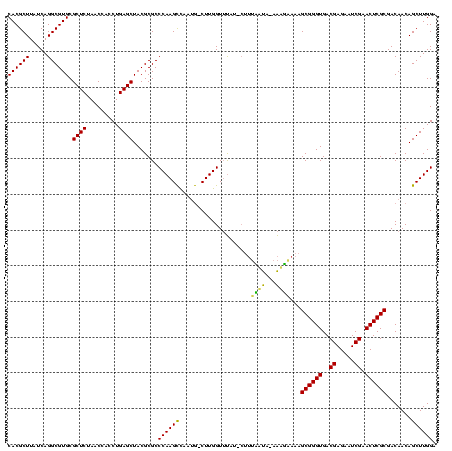
| Location | 1,839,762 – 1,839,879 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.80 |
| Mean single sequence MFE | -38.57 |
| Consensus MFE | -32.73 |
| Energy contribution | -33.07 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.51 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 1839762 117 - 2030921/80-200 GCCCAAGCCUAUG-CUUGGUUUAU-CUUUUAUA-AAAGAAAAGCGGGUGACGAGAAUCGAACUCGCGACAACAGCUUGGAAGGCUGUAGUUUUACCACUAAACUACACCCGCUUAGAUGG ..((((((....)-)))))....(-((((....-))))).(((((((((.((((.......)))).(((.((((((.....)))))).)))..............)))))))))...... ( -38.90) >sp_ag.0 1904703 119 - 2160267/80-200 GCCCAAGCUAUUG-CUUGGUUUUUACUUUCUUAUAAAGUAAAGCGGGUGACGAGAAUCGAACUCGCGACAACAGCUUGGAAGGCUGUAGUUUUACCACUAAACUACACCCGCUUCUAUGG ..((((((....)-)))))..((((((((.....))))))))(((((((.((((.......)))).(((.((((((.....)))))).)))..............)))))))........ ( -40.50) >sp_pn.0 1851494 117 + 2038615/80-200 GCCCAAGUCAAAAACUUGGUACAA---AGAACAAAGUUCAAAGCGGGUGACGAGAAUCGAACUCGCGACAACAGCUUGGAAGGCUGUAGUUUUACCACUAAACUACACCCGCUAAAAUGG ..((((((.....)))))).....---.((((...))))..((((((((.((((.......)))).(((.((((((.....)))))).)))..............))))))))....... ( -36.30) >consensus GCCCAAGCCAAUG_CUUGGUUUAU_CUUUAAUA_AAAGAAAAGCGGGUGACGAGAAUCGAACUCGCGACAACAGCUUGGAAGGCUGUAGUUUUACCACUAAACUACACCCGCUUAAAUGG ..(((((.......))))).....................(((((((((.((((.......)))).(((.((((((.....)))))).)))..............)))))))))...... (-32.73 = -33.07 + 0.33)

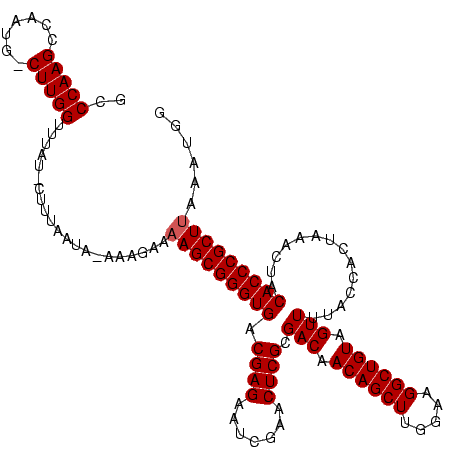
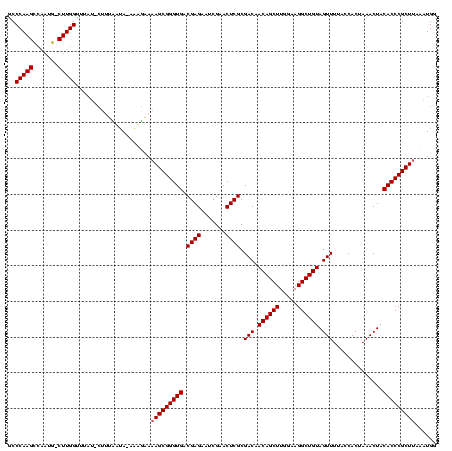
| Location | 1,839,762 – 1,839,879 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.80 |
| Mean single sequence MFE | -37.53 |
| Consensus MFE | -31.11 |
| Energy contribution | -31.00 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 1839762 117 - 2030921/80-200 CCAUCUAAGCGGGUGUAGUUUAGUGGUAAAACUACAGCCUUCCAAGCUGUUGUCGCGAGUUCGAUUCUCGUCACCCGCUUUUCUUU-UAUAAAAG-AUAAACCAAG-CAUAGGCUUGGGC ......(((((((((...............((.(((((.......))))).)).(((((.......)))))))))))))).(((((-....))))-)....(((((-(....)))))).. ( -37.10) >sp_ag.0 1904703 119 - 2160267/80-200 CCAUAGAAGCGGGUGUAGUUUAGUGGUAAAACUACAGCCUUCCAAGCUGUUGUCGCGAGUUCGAUUCUCGUCACCCGCUUUACUUUAUAAGAAAGUAAAAACCAAG-CAAUAGCUUGGGC ........((((.((((((((.......)))))))).))..((((((((((((.(((.((.((.....))..)).)))((((((((.....))))))))......)-))))))))))))) ( -41.50) >sp_pn.0 1851494 117 + 2038615/80-200 CCAUUUUAGCGGGUGUAGUUUAGUGGUAAAACUACAGCCUUCCAAGCUGUUGUCGCGAGUUCGAUUCUCGUCACCCGCUUUGAACUUUGUUCU---UUGUACCAAGUUUUUGACUUGGGC ...((..((((((((...............((.(((((.......))))).)).(((((.......)))))))))))))..))..........---.....(((((((...))))))).. ( -34.00) >consensus CCAUAUAAGCGGGUGUAGUUUAGUGGUAAAACUACAGCCUUCCAAGCUGUUGUCGCGAGUUCGAUUCUCGUCACCCGCUUUACUUU_UAUAAAAG_AUAAACCAAG_CAUUGGCUUGGGC ......(((((((((...............((.(((((.......))))).)).(((((.......)))))))))))))).....................(((((.(....)))))).. (-31.11 = -31.00 + -0.11) # Strand winner: forward (0.80)

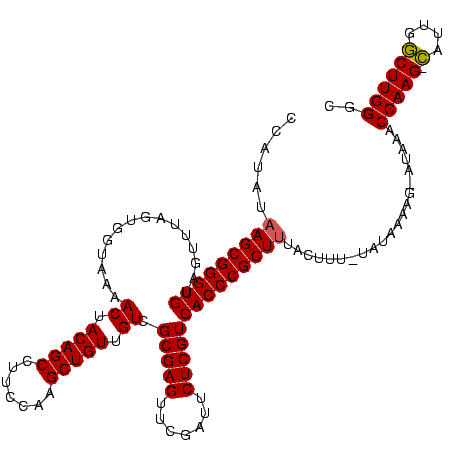
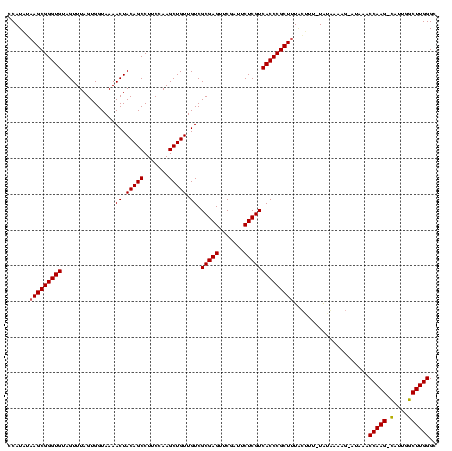
| Location | 1,839,762 – 1,839,882 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -39.07 |
| Consensus MFE | -38.40 |
| Energy contribution | -38.40 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 1839762 120 - 2030921/120-240 AAGCGGGUGACGAGAAUCGAACUCGCGACAACAGCUUGGAAGGCUGUAGUUUUACCACUAAACUACACCCGCUUAGAUGGGAGUUAACGGGCUCGAACCGCUGACCCUCUGCUUGUAAGG (((((((((.((((.......)))).(((.((((((.....)))))).)))..............)))))))))((..(((.((((.(((.......))).)))))))...))....... ( -39.60) >sp_ag.0 1904703 120 - 2160267/120-240 AAGCGGGUGACGAGAAUCGAACUCGCGACAACAGCUUGGAAGGCUGUAGUUUUACCACUAAACUACACCCGCUUCUAUGGGAGUUAACGGGAUCGAACCGCUGACCCUCUGCUUGUAAGG (((((((((.((((.......)))).(((.((((((.....)))))).)))..............)))))))))....(((.((((.(((.......))).)))))))............ ( -38.80) >sp_pn.0 1851494 120 + 2038615/120-240 AAGCGGGUGACGAGAAUCGAACUCGCGACAACAGCUUGGAAGGCUGUAGUUUUACCACUAAACUACACCCGCUAAAAUGGGAGUUAACGGGAUCGAACCGCUGACCCUCUGCUUGUAAGG .((((((((.((((.......)))).(((.((((((.....)))))).)))..............)))))))).....(((.((((.(((.......))).)))))))............ ( -38.80) >consensus AAGCGGGUGACGAGAAUCGAACUCGCGACAACAGCUUGGAAGGCUGUAGUUUUACCACUAAACUACACCCGCUUAAAUGGGAGUUAACGGGAUCGAACCGCUGACCCUCUGCUUGUAAGG .((((((((.((((.......)))).(((.((((((.....)))))).)))..............)))))))).....(((.((((.(((.......))).)))))))............ (-38.40 = -38.40 + -0.00)

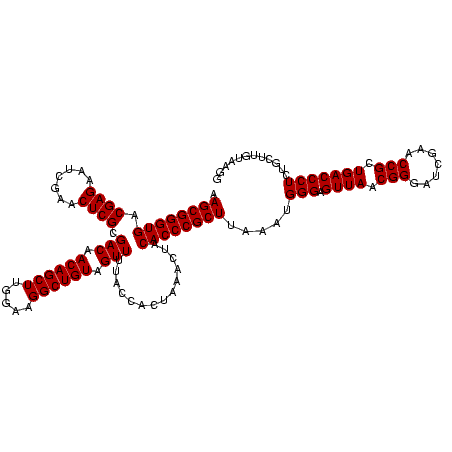
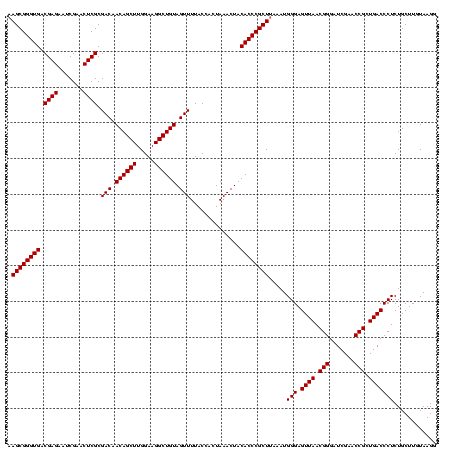
| Location | 1,839,762 – 1,839,882 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -42.90 |
| Consensus MFE | -41.17 |
| Energy contribution | -41.17 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.68 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 1839762 120 - 2030921/160-280 AGGCUGUAGUUUUACCACUAAACUACACCCGCUUAGAUGGGAGUUAACGGGCUCGAACCGCUGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAACUGAGCUAAACUCCCUCUUGCUAAG .((.((((((((.......)))))))).))((..(((.(((((((...(((.(((......))))))((((((.....)))))).((((........))))..)))))))))).)).... ( -44.30) >sp_ag.0 1904703 119 - 2160267/160-280 AGGCUGUAGUUUUACCACUAAACUACACCCGCUUCUAUGGGAGUUAACGGGAUCGAACCGCUGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAGCUGAGCUAAACUCCCU-UUGCUAAG .((.((((((((.......)))))))).))((......(((((((...(((.(((......))))))((((((.....)))))).((((........))))..))))))).-..)).... ( -41.00) >sp_pn.0 1851494 120 + 2038615/160-280 AGGCUGUAGUUUUACCACUAAACUACACCCGCUAAAAUGGGAGUUAACGGGAUCGAACCGCUGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAGCUGAGCUAAACUCCCUAGAGCUAAG .((.((((((((.......)))))))).))(((.....(((((((...(((.(((......))))))((((((.....)))))).((((........))))..)))))))...))).... ( -43.40) >consensus AGGCUGUAGUUUUACCACUAAACUACACCCGCUUAAAUGGGAGUUAACGGGAUCGAACCGCUGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAGCUGAGCUAAACUCCCU_UUGCUAAG .((.((((((((.......)))))))).))((......(((((((...(((.(((......))))))((((((.....)))))).((((........))))..)))))))....)).... (-41.17 = -41.17 + -0.00)

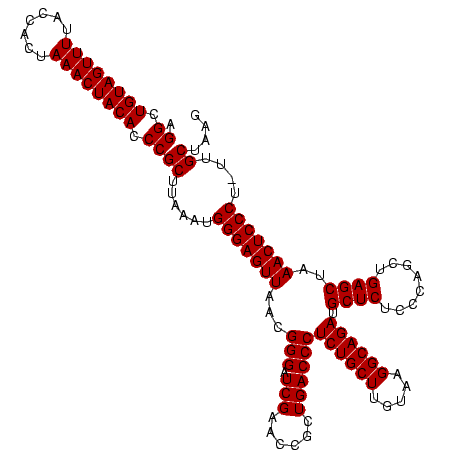
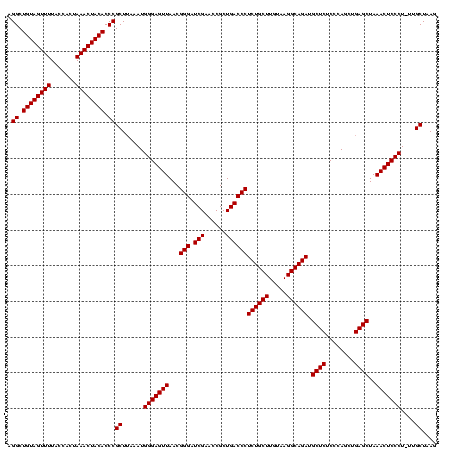
| Location | 1,839,762 – 1,839,882 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -41.60 |
| Consensus MFE | -39.60 |
| Energy contribution | -39.60 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 1839762 120 - 2030921/160-280 CUUAGCAAGAGGGAGUUUAGCUCAGUUGGGAGAGCAUCUGCCUUACAAGCAGAGGGUCAGCGGUUCGAGCCCGUUAACUCCCAUCUAAGCGGGUGUAGUUUAGUGGUAAAACUACAGCCU ....((.((((((((((..((((........)))).(((((.......)))))((((...((...)).))))...))))))).)))..))((.((((((((.......)))))))).)). ( -42.80) >sp_ag.0 1904703 119 - 2160267/160-280 CUUAGCAA-AGGGAGUUUAGCUCAGCUGGGAGAGCAUCUGCCUUACAAGCAGAGGGUCAGCGGUUCGAUCCCGUUAACUCCCAUAGAAGCGGGUGUAGUUUAGUGGUAAAACUACAGCCU ........-.(((((((.(((...)))((((((((..(((((((........)))).)))..))))..))))...)))))))........((.((((((((.......)))))))).)). ( -40.10) >sp_pn.0 1851494 120 + 2038615/160-280 CUUAGCUCUAGGGAGUUUAGCUCAGCUGGGAGAGCAUCUGCCUUACAAGCAGAGGGUCAGCGGUUCGAUCCCGUUAACUCCCAUUUUAGCGGGUGUAGUUUAGUGGUAAAACUACAGCCU ...(((((....)))))..(((.((.(((((((((.(((((.......)))))(((((........))))).)))..)))))).)).)))((.((((((((.......)))))))).)). ( -41.90) >consensus CUUAGCAA_AGGGAGUUUAGCUCAGCUGGGAGAGCAUCUGCCUUACAAGCAGAGGGUCAGCGGUUCGAUCCCGUUAACUCCCAUAUAAGCGGGUGUAGUUUAGUGGUAAAACUACAGCCU ..........(((((((..((((........)))).(((((.......)))))(((((........)).)))...)))))))........((.((((((((.......)))))))).)). (-39.60 = -39.60 + 0.00) # Strand winner: reverse (0.92)

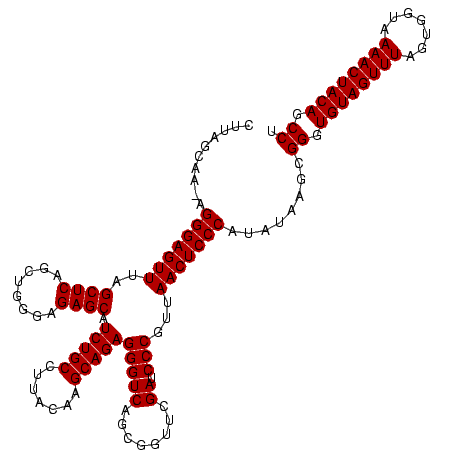
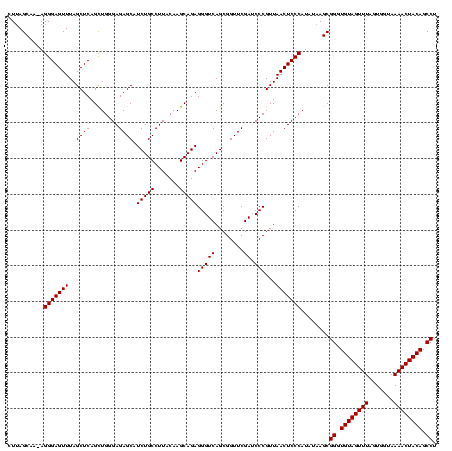
| Location | 1,839,762 – 1,839,882 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -36.13 |
| Consensus MFE | -33.46 |
| Energy contribution | -32.80 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.822154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_mu.0 1839762 120 - 2030921/200-320 GAGUUAACGGGCUCGAACCGCUGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAACUGAGCUAAACUCCCUCUUGCUAAGCGACGACCCUAUCUCACAGGGGGCAACCCCCAACUACUUC (((.....(((.(((...((((.....((((((.....)))))).((((........)))).................)))).))))))...)))...((((....)))).......... ( -37.50) >sp_ag.0 1904703 119 - 2160267/200-320 GAGUUAACGGGAUCGAACCGCUGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAGCUGAGCUAAACUCCCU-UUGCUAAGCGACUACCUUAUCUCACAGGGGGCAACCCCCAACUACUUC (((.(((.(((.(((......))))))..(((((((((((.((..((((........))))....)).)).-)))).)))))......))).)))...((((....)))).......... ( -34.00) >sp_pn.0 1851494 120 + 2038615/200-320 GAGUUAACGGGAUCGAACCGCUGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAGCUGAGCUAAACUCCCUAGAGCUAAGCGACUUCCCUAUCUCACAGGGGGCAACCCCCAACUACUUC (((.....(((((((....(((.....((((((.....)))))).((((........))))............)))....)))..))))...)))...((((....)))).......... ( -36.90) >consensus GAGUUAACGGGAUCGAACCGCUGACCCUCUGCUUGUAAGGCAGAUGCUCUCCCAGCUGAGCUAAACUCCCU_UUGCUAAGCGACUACCCUAUCUCACAGGGGGCAACCCCCAACUACUUC (((.....(((((((....((......((((((.....)))))).((((........)))).............))....))).).)))...)))...((((....)))).......... (-33.46 = -32.80 + -0.66)

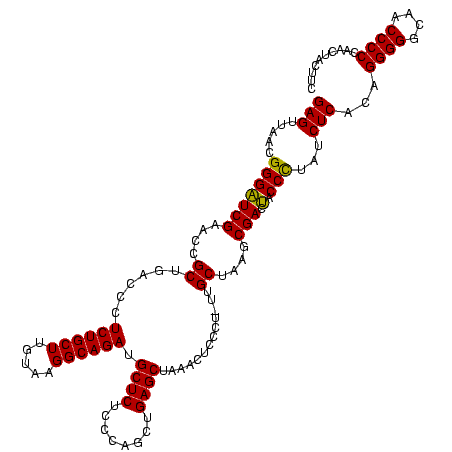
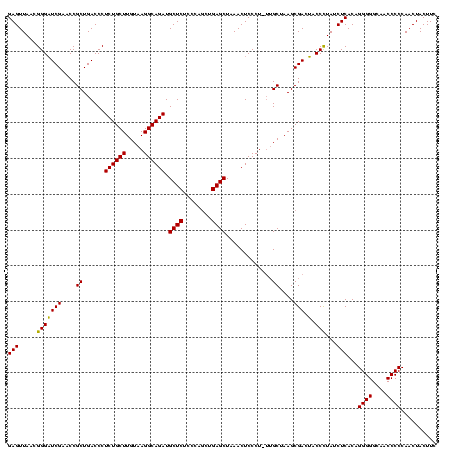
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:51:15 2006