
| Sequence ID | sy_sa.0 |
|---|---|
| Location | 1,425,087 – 1,425,207 |
| Length | 120 |
| Max. P | 0.999745 |

| Location | 1,425,087 – 1,425,205 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.46 |
| Mean single sequence MFE | -22.47 |
| Consensus MFE | -15.19 |
| Energy contribution | -15.30 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.68 |
| SVM decision value | 3.99 |
| SVM RNA-class probability | 0.999745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
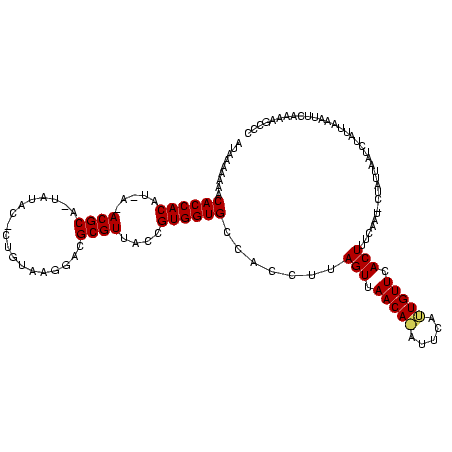
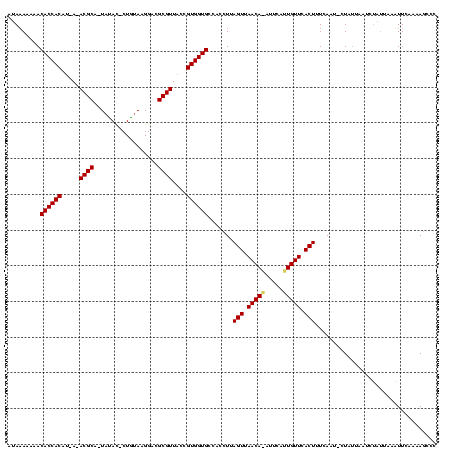
>sy_sa.0 1425087 118 + 2516575/0-120 AUAAAAAAACACCACAUUA-ACGCA-UACAUACUGUAAGGACGCGUUACCGUGGUGCCACCUUAGUUAACAAAUUCAUUGUUCACUUUCAUUGCUAUUAAUUUAGUACAUUUAAAAGCCC .........((((((..((-((((.-(((.....))).....))))))..)))))).......(((.(((((.....))))).))).....(((((......)))))............. ( -21.90) >sy_au.0 1395440 114 - 2809422/0-120 AUAAAAAGACACCACAU-A-ACGCA-UAUAC-CUGUAAGGACGCGUUACCGUGGUGCCACCUUAGUUAACAUAAUUCAUGUUCACUUUCAAAACU--CAUUCAAUUGUUUUCCAAAGUCC .......(.((((((.(-(-((((.-....(-(.....))..))))))..)))))).).....(((.(((((.....))))).)))...(((((.--.........)))))......... ( -20.50) >sy_ha.0 1583194 117 + 2685015/0-120 ACAUAAAAACACCACAGCAUACGCGCUUUACUCUAUAAGGACGCGUUACCGUGGUGCCACCUUAGUUAACA-AUACUUUGUUCACUCUUACU-UUAUUAAAGUUUAAAAUUCUAGA-CCC .........((((((.(...((((((((((.....))))).)))))..).)))))).......(((.((((-(....))))).)))......-........(((((......))))-).. ( -25.00) >consensus AUAAAAAAACACCACAU_A_ACGCA_UAUAC_CUGUAAGGACGCGUUACCGUGGUGCCACCUUAGUUAACA_AUUCAUUGUUCACUUUCAAU_CUAUUAAUCUAUUAAAUUCAAAAGCCC .........((((((.....((((..................))))....)))))).......(((.(((((.....))))).))).................................. (-15.19 = -15.30 + 0.11)

| Location | 1,425,087 – 1,425,205 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.46 |
| Mean single sequence MFE | -31.73 |
| Consensus MFE | -21.62 |
| Energy contribution | -21.29 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.68 |
| SVM decision value | 3.78 |
| SVM RNA-class probability | 0.999607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
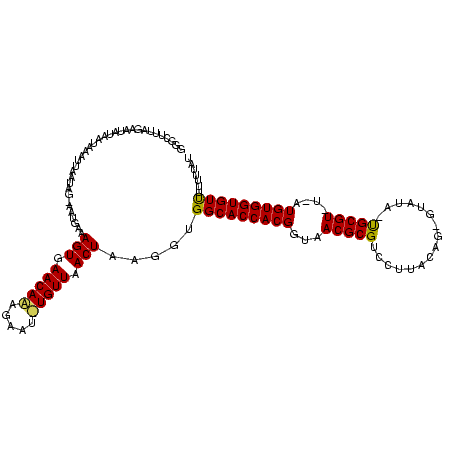
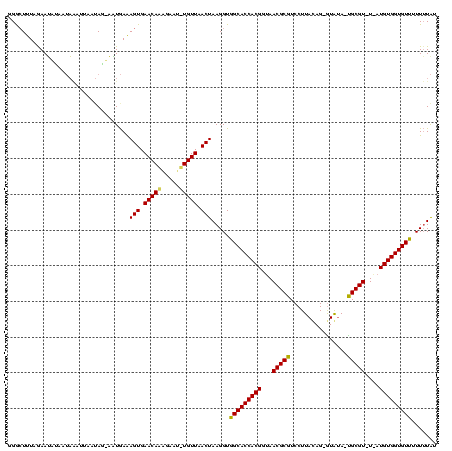
>sy_sa.0 1425087 118 + 2516575/0-120 GGGCUUUUAAAUGUACUAAAUUAAUAGCAAUGAAAGUGAACAAUGAAUUUGUUAACUAAGGUGGCACCACGGUAACGCGUCCUUACAGUAUGUA-UGCGU-UAAUGUGGUGUUUUUUUAU ..(((.((((.(......).)))).)))......(((.(((((.....))))).)))((((..((((((((.((((((((.(.........).)-)))))-)).))))))))..)))).. ( -31.10) >sy_au.0 1395440 114 - 2809422/0-120 GGACUUUGGAAAACAAUUGAAUG--AGUUUUGAAAGUGAACAUGAAUUAUGUUAACUAAGGUGGCACCACGGUAACGCGUCCUUACAG-GUAUA-UGCGU-U-AUGUGGUGUCUUUUUAU ..((((((((((((.........--.))))).......((((((...))))))..)))))))(((((((((.((((((((((.....)-)...)-)))))-)-))))))))))....... ( -33.50) >sy_ha.0 1583194 117 + 2685015/0-120 GGG-UCUAGAAUUUUAAACUUUAAUAA-AGUAAGAGUGAACAAAGUAU-UGUUAACUAAGGUGGCACCACGGUAACGCGUCCUUAUAGAGUAAAGCGCGUAUGCUGUGGUGUUUUUAUGU ...-...................((((-((....(((.(((((....)-)))).)))......(((((((((((((((((..((((...)))).)))))).))))))))))))))))).. ( -30.60) >consensus GGGCUUUAGAAUAUAAUAAAUUAAUAG_AAUGAAAGUGAACAAAGAAU_UGUUAACUAAGGUGGCACCACGGUAACGCGUCCUUACAG_GUAUA_UGCGU_U_AUGUGGUGUUUUUUUAU ..................................(((.(((((.....))))).))).....(((((((((...(((((................)))))....)))))))))....... (-21.62 = -21.29 + -0.33) # Strand winner: reverse (1.00)

| Location | 1,425,087 – 1,425,207 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.50 |
| Mean single sequence MFE | -21.66 |
| Consensus MFE | -17.82 |
| Energy contribution | -15.39 |
| Covariance contribution | -2.43 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993499 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 1425087 120 + 2516575/86-206 AUUUGAAAGAUAACGUAGCAAAUAUGUCAUGUAGCAAAAAGAAAGCUAAUGGUUGAUGAAAAUUAGCACUGCAUAUAUUGUGAAUUGGGCUUUUAAAUGUACUAAAUUAAUAGCAAUGAA (((((((((........(((((((((((((.((((.........)))))))((((((....))))))...)))))).))))........)))))))))...................... ( -22.79) >sy_au.0 1395440 118 - 2809422/86-206 UUUCUAAAGAAAUAGUAGCAGAUAUGAAACGUAGCAAAUAGAAAGCUAAUGGGUGAUGGGAAUUAGCACGCCAUAUCUUGUGAAUUGGACUUUGGAAAACAAUUGAAUG--AGUUUUGAA (((((((((...((((.((((((((....((((((.........))).)))((((.((........))))))))))).)))..))))..)))))))))...........--......... ( -22.70) >sy_ha.0 1583194 118 + 2685015/86-206 AUUCUAUGAAUGAAGUAGAUAAUAUGAUUUGUAGCAAAUAGAAAGCCAAUGGGUGAUGUAAAUUGGCACUACAUAUUAAUCGAAUUGGG-UCUAGAAUUUUAAACUUUAAUAA-AGUAAG ((((((.((.(.((...((((((((((((((...))))).....((((((...........))))))....)))))).)))...)).).-)))))))).....((((.....)-)))... ( -19.50) >consensus AUUCUAAAGAUAAAGUAGCAAAUAUGAAAUGUAGCAAAUAGAAAGCUAAUGGGUGAUGAAAAUUAGCACUACAUAUAUUGUGAAUUGGGCUUUAGAAUAUAAUAAAUUAAUAG_AAUGAA (((((((((........(((((((((........(.....)...((((((...........))))))....)))))).)))........)))))))))...................... (-17.82 = -15.39 + -2.43) # Strand winner: reverse (1.00)

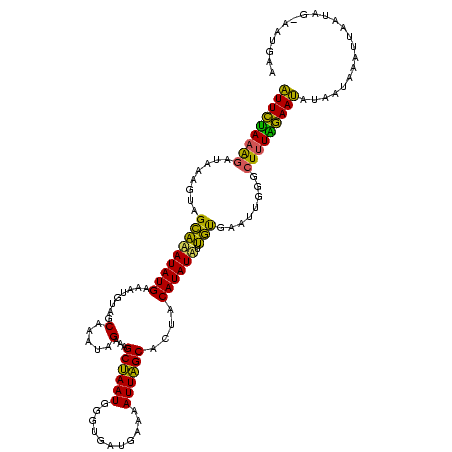
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:53:36 2006