| Sequence ID | sy_ha.0 |
|---|---|
| Location | 1,408,601 – 1,408,716 |
| Length | 115 |
| Max. P | 0.997998 |

| Location | 1,408,601 – 1,408,716 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.44 |
| Mean single sequence MFE | -30.17 |
| Consensus MFE | -26.64 |
| Energy contribution | -26.09 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
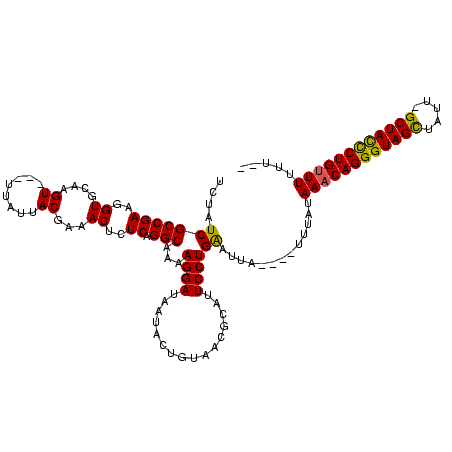
>sy_ha.0 1408601 115 + 2685015/0-120 UUUAUCGCCGAAGGUGCAAGU---UAACUACUAAACUCUCAGGCAAAAGGAUAAUACUGCUACGCAUUCCUGGAUUA--AGUGUAUAAACAGGGUAGCUAUUUGCUACUCUGUUUUUUAU ...((((((((..((...(((---.....)))..))..)).)))...((((......(((...))).)))).)))..--.(((...((((((((((((.....))))))))))))..))) ( -29.80) >sy_au.0 1179415 116 - 2809422/0-120 UCUAUCGCCGAAGGUGCAAGUAAUUUAUUACGAAACUCUCAGGCAAAAGGAUAAUACUGUAACGCGUUCCUGAAUUGGUGAUUUAUAAACAGGGUAGCGAUU-GCUAUCCUGUUUUU--- ...((((((((.(((....((((....))))...))).(((((.((..(.............)...))))))).))))))))....((((((((((((....-))))))))))))..--- ( -32.42) >sy_sa.0 1259195 109 + 2516575/0-120 UCCAUCGCCGAAGGUGCAAGU----UUUUACGAAACUCUCAGGCAAAAGGAUAAUACUGUAACGCAUUCCUGAAAUA-----UUACAA-CAGGGUAGUU-UAGGCUACCCUGUUUUUUUA ....(((((((..((....((----....))...))..)).)))...((((................))))))....-----....((-(((((((((.-...)))))))))))...... ( -28.29) >consensus UCUAUCGCCGAAGGUGCAAGU___UUAUUACGAAACUCUCAGGCAAAAGGAUAAUACUGUAACGCAUUCCUGAAUUA____UUUAUAAACAGGGUAGCUAUU_GCUACCCUGUUUUUU__ ....(((((((..((....((........))...))..)).)))...((((................)))))).............((((((((((((.....))))))))))))..... (-26.64 = -26.09 + -0.55)

| Location | 1,408,601 – 1,408,716 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.44 |
| Mean single sequence MFE | -28.43 |
| Consensus MFE | -25.22 |
| Energy contribution | -26.67 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
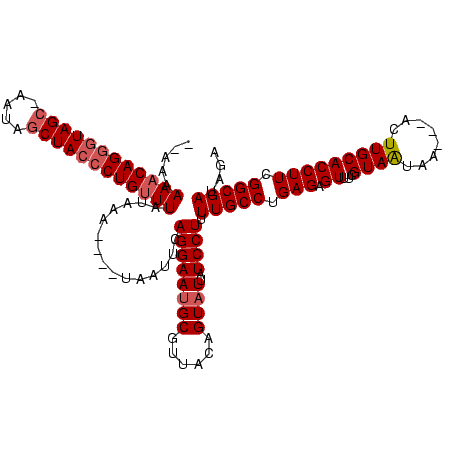
>sy_ha.0 1408601 115 + 2685015/0-120 AUAAAAAACAGAGUAGCAAAUAGCUACCCUGUUUAUACACU--UAAUCCAGGAAUGCGUAGCAGUAUUAUCCUUUUGCCUGAGAGUUUAGUAGUUA---ACUUGCACCUUCGGCGAUAAA .....((((((.(((((.....))))).)))))).......--......((((((((......))))..)))).(((((.(((.((..(((.....---)))...))))).))))).... ( -28.50) >sy_au.0 1179415 116 - 2809422/0-120 ---AAAAACAGGAUAGC-AAUCGCUACCCUGUUUAUAAAUCACCAAUUCAGGAACGCGUUACAGUAUUAUCCUUUUGCCUGAGAGUUUCGUAAUAAAUUACUUGCACCUUCGGCGAUAGA ---..(((((((.((((-....)))).)))))))....(((........((((..((......))....))))...(((.(((.((...((((....))))....))))).))))))... ( -25.70) >sy_sa.0 1259195 109 + 2516575/0-120 UAAAAAAACAGGGUAGCCUA-AACUACCCUG-UUGUAA-----UAUUUCAGGAAUGCGUUACAGUAUUAUCCUUUUGCCUGAGAGUUUCGUAAAA----ACUUGCACCUUCGGCGAUGGA ......((((((((((....-..))))))))-))....-----.........(((((......))))).(((..(((((.(((.((...((((..----..))))))))).))))).))) ( -31.10) >consensus __AAAAAACAGGGUAGC_AAUAGCUACCCUGUUUAUAAA____UAAUUCAGGAAUGCGUUACAGUAUUAUCCUUUUGCCUGAGAGUUUCGUAAUAA___ACUUGCACCUUCGGCGAUAGA .....((((((((((((.....))))))))))))...............((((((((......))))..)))).(((((.(((.((...((((........))))))))).))))).... (-25.22 = -26.67 + 1.45) # Strand winner: forward (0.94)

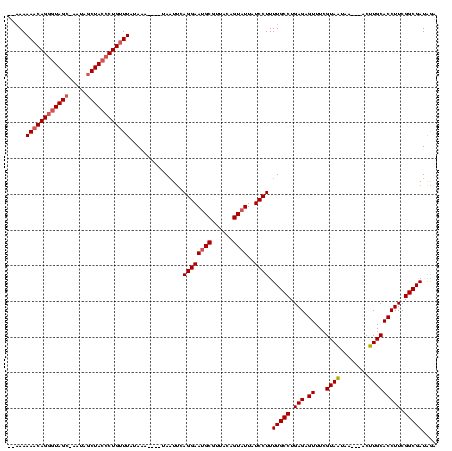
| Location | 1,408,601 – 1,408,713 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.43 |
| Mean single sequence MFE | -25.53 |
| Consensus MFE | -21.99 |
| Energy contribution | -21.22 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.993923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1408601 112 + 2685015/18-138 AGU---UAACUACUAAACUCUCAGGCAAAAGGAUAAUACUGCUACGCAUUCCUGGAUUA--AGUGUAUAAACAGGGUAGCUAUUUGCUACUCUGUUUUUUAUA---AAAAUUAUAGGGGG ...---...........(((((.......((((......(((...))).)))).((((.--..((((.((((((((((((.....))))))))))))..))))---..))))...))))) ( -27.20) >sy_au.0 1179415 112 - 2809422/18-138 AGUAAUUUAUUACGAAACUCUCAGGCAAAAGGAUAAUACUGUAACGCGUUCCUGAAUUGGUGAUUUAUAAACAGGGUAGCGAUU-GCUAUCCUGUUUUU-------AUAAUUUUAAGGGG .((((....))))....((((((((.((..(.............)...)))))))......((((...((((((((((((....-))))))))))))..-------..))))....))). ( -24.92) >sy_sa.0 1259195 109 + 2516575/18-138 AGU----UUUUACGAAACUCUCAGGCAAAAGGAUAAUACUGUAACGCAUUCCUGAAAUA-----UUACAA-CAGGGUAGUU-UAGGCUACCCUGUUUUUUUAUUAUAUAAUCUUUAGGAG ...----..........((((((((.......(((....)))........)))))((((-----..(.((-(((((((((.-...))))))))))).)..)))).............))) ( -24.46) >consensus AGU___UUAUUACGAAACUCUCAGGCAAAAGGAUAAUACUGUAACGCAUUCCUGAAUUA____UUUAUAAACAGGGUAGCUAUU_GCUACCCUGUUUUUU______AUAAUUUUAAGGGG .................((((((((.....(.............).....))))).............((((((((((((.....))))))))))))....................))) (-21.99 = -21.22 + -0.77)

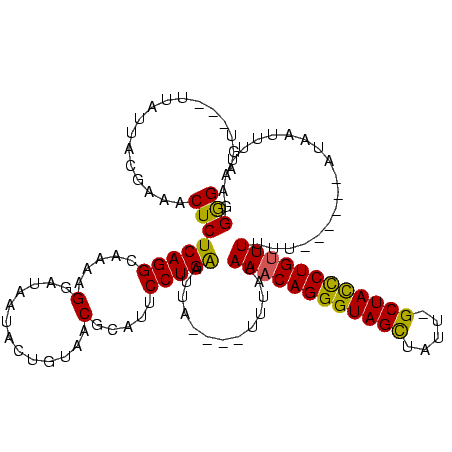
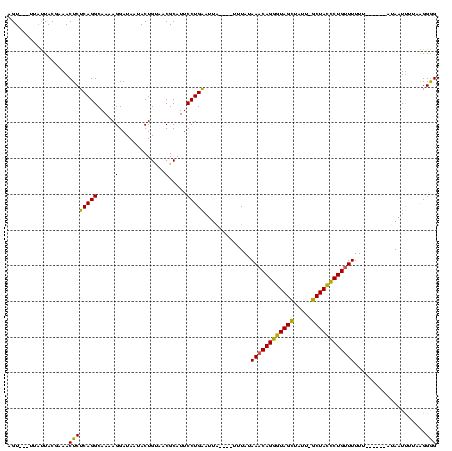
| Location | 1,408,601 – 1,408,713 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.43 |
| Mean single sequence MFE | -22.87 |
| Consensus MFE | -17.37 |
| Energy contribution | -19.37 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.76 |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.997998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1408601 112 + 2685015/18-138 CCCCCUAUAAUUUU---UAUAAAAAACAGAGUAGCAAAUAGCUACCCUGUUUAUACACU--UAAUCCAGGAAUGCGUAGCAGUAUUAUCCUUUUGCCUGAGAGUUUAGUAGUUA---ACU ....((((((((((---((....((((((.(((((.....))))).)))))).......--......((((((((......))))..))))......))))))))..))))...---... ( -23.20) >sy_au.0 1179415 112 - 2809422/18-138 CCCCUUAAAAUUAU-------AAAAACAGGAUAGC-AAUCGCUACCCUGUUUAUAAAUCACCAAUUCAGGAACGCGUUACAGUAUUAUCCUUUUGCCUGAGAGUUUCGUAAUAAAUUACU .........(((((-------..(((((((.((((-....)))).)))))))................(((((((......))......(((......))).))))))))))........ ( -19.80) >sy_sa.0 1259195 109 + 2516575/18-138 CUCCUAAAGAUUAUAUAAUAAAAAAACAGGGUAGCCUA-AACUACCCUG-UUGUAA-----UAUUUCAGGAAUGCGUUACAGUAUUAUCCUUUUGCCUGAGAGUUUCGUAAAA----ACU ..........((((..(((.....((((((((((....-..))))))))-))....-----..((((((((((((......))))).........))))))))))..))))..----... ( -25.60) >consensus CCCCUUAAAAUUAU______AAAAAACAGGGUAGC_AAUAGCUACCCUGUUUAUAAA____UAAUUCAGGAAUGCGUUACAGUAUUAUCCUUUUGCCUGAGAGUUUCGUAAUAA___ACU .......................((((((((((((.....))))))))))))............(((((((((((......))))).........))))))................... (-17.37 = -19.37 + 2.00) # Strand winner: forward (1.00)

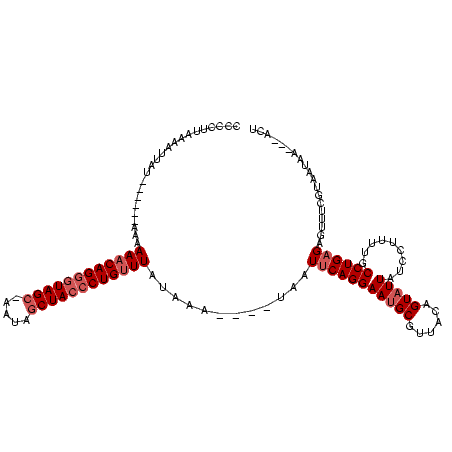
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:53:33 2006