
| Sequence ID | sy_au.0 |
|---|---|
| Location | 1,974,471 – 1,974,570 |
| Length | 99 |
| Max. P | 0.997329 |

| Location | 1,974,471 – 1,974,570 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 83.08 |
| Mean single sequence MFE | -36.12 |
| Consensus MFE | -34.75 |
| Energy contribution | -32.75 |
| Covariance contribution | -2.00 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
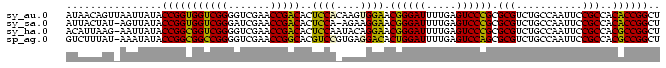
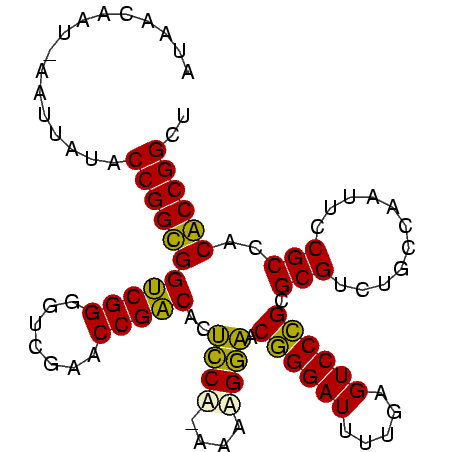
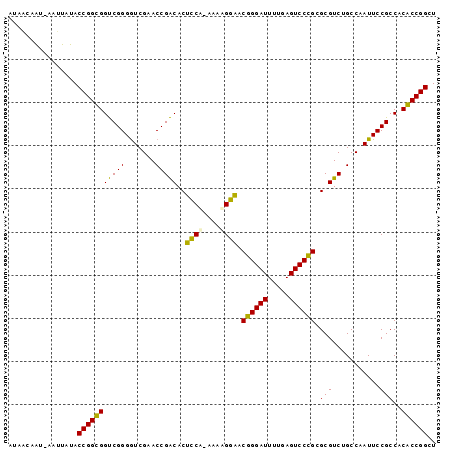
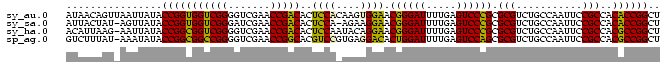
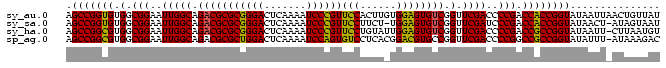
>sy_au.0 1974471 99 + 2809422 AUAACAGUUAAUUAUACCGGUGGUCGGGGUCGAACCGACACUCCACAAGUGGAACGGGAUUUUGAGUCCCGCGCGUCUGCCAAUUCCGCCACACCGGCU ................(((((((((((.......))))).........((((((((((((.....)))))).((....))...))))))..)))))).. ( -35.80) >sy_sa.0 1548537 97 - 2516575 AUUACUAU-AGUUAUACCGGUGGUCGGGAUCGAACCGACACUCCA-AGAAGGAACGGGAUUUUGAGUCCCGCGCGUCUGCCAAUUCCGCCACACCGGCU ........-.......(((((((((((.......)))))......-....((((((((((.....)))))).((....))...))))....)))))).. ( -32.80) >sy_ha.0 1574106 98 - 2685015 ACAUUAAG-AAUUAUACCGGCGGUCGGGGUCGAACCGACACUCCAAUACAGGAACGGGAUUUUGAGUCCCGCGCGUCUGCCAAUUCCGCCACGCCGGCU ........-.......(((((((((((.......)))))...........((((((((((.....)))))).((....))...))))....)))))).. ( -34.30) >sp_ag.0 2062846 98 - 2160267 GUCUUUAU-AAAUAUACCGGCGGCCGGGGUCGAACCGGCACGUCCGUGAGGACACUGGAUUUUGAGUCCAGCGCGUCUGCCAAUUCCGCCACGCCGGCU ........-.......(((((((.(((((.....))((((.((((....)))).((((((.....))))))......))))....))).).)))))).. ( -41.60) >consensus AUAACAAU_AAUUAUACCGGCGGUCGGGGUCGAACCGACACUCCA_AAAAGGAACGGGAUUUUGAGUCCCGCGCGUCUGCCAAUUCCGCCACACCGGCU ................(((((((((((.......)))))..(((......))).((((((.....)))))).(((...........)))..)))))).. (-34.75 = -32.75 + -2.00)
| Location | 1,974,471 – 1,974,570 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 83.08 |
| Mean single sequence MFE | -40.52 |
| Consensus MFE | -40.10 |
| Energy contribution | -38.72 |
| Covariance contribution | -1.37 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
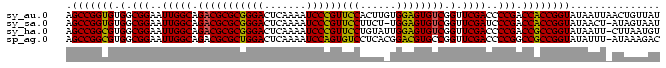
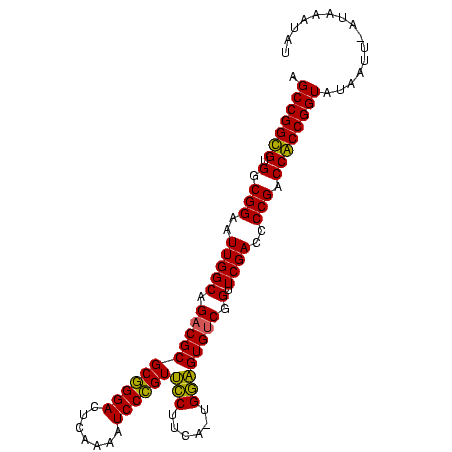
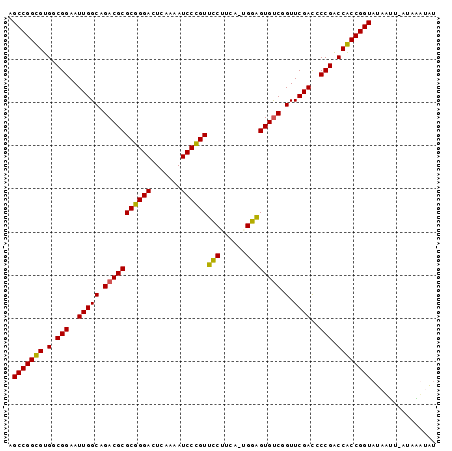
>sy_au.0 1974471 99 + 2809422 AGCCGGUGUGGCGGAAUUGGCAGACGCGCGGGACUCAAAAUCCCGUUCCACUUGUGGAGUGUCGGUUCGACCCCGACCACCGGUAUAAUUAACUGUUAU .(((((((.(.(((..(((((.(((((((((((.......))))))((((....))))))))).).))))..))).))))))))............... ( -40.00) >sy_sa.0 1548537 97 - 2516575 AGCCGGUGUGGCGGAAUUGGCAGACGCGCGGGACUCAAAAUCCCGUUCCUUCU-UGGAGUGUCGGUUCGAUCCCGACCACCGGUAUAACU-AUAGUAAU .(((((((.(.(((.((((((.(((((((((((.......))))))(((....-.)))))))).).))))).))).))))))))......-........ ( -40.00) >sy_ha.0 1574106 98 - 2685015 AGCCGGCGUGGCGGAAUUGGCAGACGCGCGGGACUCAAAAUCCCGUUCCUGUAUUGGAGUGUCGGUUCGACCCCGACCGCCGGUAUAAUU-CUUAAUGU .(((((((.(.(((..(((((.(((((((((((.......))))))(((......)))))))).).))))..))).))))))))......-........ ( -40.10) >sp_ag.0 2062846 98 - 2160267 AGCCGGCGUGGCGGAAUUGGCAGACGCGCUGGACUCAAAAUCCAGUGUCCUCACGGACGUGCCGGUUCGACCCCGGCCGCCGGUAUAUUU-AUAAAGAC .(((((((.(.(((((((((((.....((((((.......))))))((((....)))).)))))))).....))).))))))))......-........ ( -42.00) >consensus AGCCGGCGUGGCGGAAUUGGCAGACGCGCGGGACUCAAAAUCCCGUUCCUUCA_UGGAGUGUCGGUUCGACCCCGACCACCGGUAUAAUU_AUAAAUAU .(((((((.(.(((..(((((.(((((((((((.......))))))(((......)))))))).).))))..))).))))))))............... (-40.10 = -38.72 + -1.37) # Strand winner: reverse (1.00)
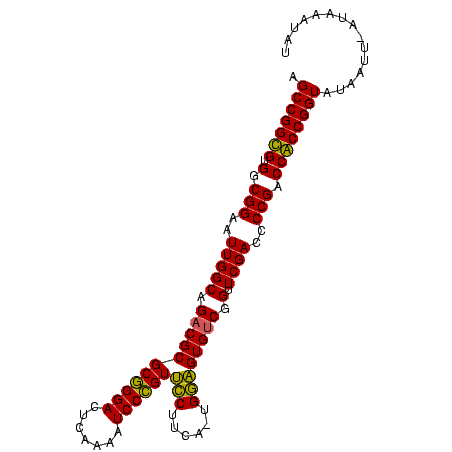
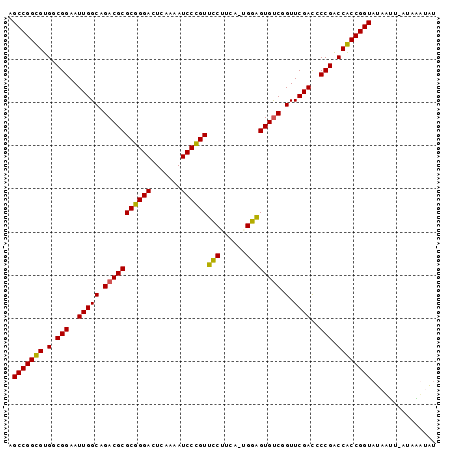
| Location | 1,974,471 – 1,974,570 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 83.08 |
| Mean single sequence MFE | -36.12 |
| Consensus MFE | -34.75 |
| Energy contribution | -32.75 |
| Covariance contribution | -2.00 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1974471 99 + 2809422/0-99 AUAACAGUUAAUUAUACCGGUGGUCGGGGUCGAACCGACACUCCACAAGUGGAACGGGAUUUUGAGUCCCGCGCGUCUGCCAAUUCCGCCACACCGGCU ................(((((((((((.......))))).........((((((((((((.....)))))).((....))...))))))..)))))).. ( -35.80) >sy_sa.0 1548537 97 - 2516575/0-99 AUUACUAU-AGUUAUACCGGUGGUCGGGAUCGAACCGACACUCCA-AGAAGGAACGGGAUUUUGAGUCCCGCGCGUCUGCCAAUUCCGCCACACCGGCU ........-.......(((((((((((.......)))))......-....((((((((((.....)))))).((....))...))))....)))))).. ( -32.80) >sy_ha.0 1574106 98 - 2685015/0-99 ACAUUAAG-AAUUAUACCGGCGGUCGGGGUCGAACCGACACUCCAAUACAGGAACGGGAUUUUGAGUCCCGCGCGUCUGCCAAUUCCGCCACGCCGGCU ........-.......(((((((((((.......)))))...........((((((((((.....)))))).((....))...))))....)))))).. ( -34.30) >sp_ag.0 2062846 98 - 2160267/0-99 GUCUUUAU-AAAUAUACCGGCGGCCGGGGUCGAACCGGCACGUCCGUGAGGACACUGGAUUUUGAGUCCAGCGCGUCUGCCAAUUCCGCCACGCCGGCU ........-.......(((((((.(((((.....))((((.((((....)))).((((((.....))))))......))))....))).).)))))).. ( -41.60) >consensus AUAACAAU_AAUUAUACCGGCGGUCGGGGUCGAACCGACACUCCA_AAAAGGAACGGGAUUUUGAGUCCCGCGCGUCUGCCAAUUCCGCCACACCGGCU ................(((((((((((.......)))))..(((......))).((((((.....)))))).(((...........)))..)))))).. (-34.75 = -32.75 + -2.00)
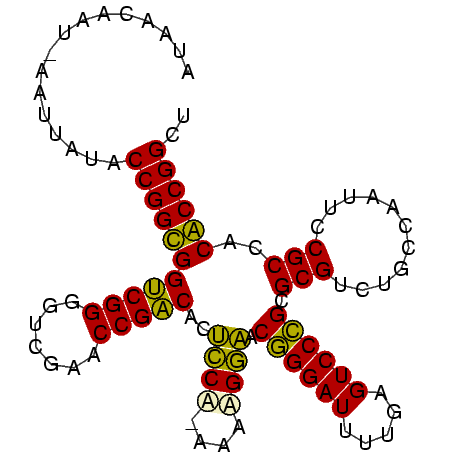

| Location | 1,974,471 – 1,974,570 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 83.08 |
| Mean single sequence MFE | -40.52 |
| Consensus MFE | -40.10 |
| Energy contribution | -38.72 |
| Covariance contribution | -1.37 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1974471 99 + 2809422/0-99 AGCCGGUGUGGCGGAAUUGGCAGACGCGCGGGACUCAAAAUCCCGUUCCACUUGUGGAGUGUCGGUUCGACCCCGACCACCGGUAUAAUUAACUGUUAU .(((((((.(.(((..(((((.(((((((((((.......))))))((((....))))))))).).))))..))).))))))))............... ( -40.00) >sy_sa.0 1548537 97 - 2516575/0-99 AGCCGGUGUGGCGGAAUUGGCAGACGCGCGGGACUCAAAAUCCCGUUCCUUCU-UGGAGUGUCGGUUCGAUCCCGACCACCGGUAUAACU-AUAGUAAU .(((((((.(.(((.((((((.(((((((((((.......))))))(((....-.)))))))).).))))).))).))))))))......-........ ( -40.00) >sy_ha.0 1574106 98 - 2685015/0-99 AGCCGGCGUGGCGGAAUUGGCAGACGCGCGGGACUCAAAAUCCCGUUCCUGUAUUGGAGUGUCGGUUCGACCCCGACCGCCGGUAUAAUU-CUUAAUGU .(((((((.(.(((..(((((.(((((((((((.......))))))(((......)))))))).).))))..))).))))))))......-........ ( -40.10) >sp_ag.0 2062846 98 - 2160267/0-99 AGCCGGCGUGGCGGAAUUGGCAGACGCGCUGGACUCAAAAUCCAGUGUCCUCACGGACGUGCCGGUUCGACCCCGGCCGCCGGUAUAUUU-AUAAAGAC .(((((((.(.(((((((((((.....((((((.......))))))((((....)))).)))))))).....))).))))))))......-........ ( -42.00) >consensus AGCCGGCGUGGCGGAAUUGGCAGACGCGCGGGACUCAAAAUCCCGUUCCUUCA_UGGAGUGUCGGUUCGACCCCGACCACCGGUAUAAUU_AUAAAUAU .(((((((.(.(((..(((((.(((((((((((.......))))))(((......)))))))).).))))..))).))))))))............... (-40.10 = -38.72 + -1.37) # Strand winner: reverse (1.00)
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:53:28 2006