| Sequence ID | sy_ha.0 |
|---|---|
| Location | 1,337,670 – 1,337,789 |
| Length | 119 |
| Max. P | 0.999967 |

| Location | 1,337,670 – 1,337,789 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.39 |
| Mean single sequence MFE | -21.90 |
| Consensus MFE | -18.97 |
| Energy contribution | -16.43 |
| Covariance contribution | -2.53 |
| Combinations/Pair | 1.55 |
| Mean z-score | -0.84 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1337670 119 + 2685015/0-120 GAAAAUACUUAAAAUAAUCUUAAGUUCGCAUGAAGAUGAGUAGGCAAUUAUUACUUAUAUAGAGAUGUACUGGUUGGUGAAAAGUGCAAUGUAUAUUGUUGAACGCUACUUCAUGU-CAU ......((((((.......))))))..((((((((.(((((((.......)))))))((((.(..((((((...........)))))).).)))).............))))))))-... ( -22.50) >sy_au.0 1107241 120 - 2809422/0-120 AAAUGUAACUAAAAUAGAAUUAAAUUCGUAUAAAGAUGAGUAGUUAAUGAUUUGUUUAUCAGAGAUGUACUGGUUGGUGAAAAGUACAACAAACAUUAGUGAACGCUACUUUAUGCACAA ...(((..........((((...))))((((((((...(((...(((((.(((((..........((((((...........))))))))))))))))(....)))).))))))))))). ( -19.80) >sy_sa.0 1191327 119 + 2516575/0-120 GAAAAUAAAUACAAUAUAAUUAACUUCACAUAAAGAUGAGUAAAUUAU-ACUGAGCGAAAAGAGAUGUACUGGUUGGUGAAAAGUACAGUCGGUAUAGUUGAAUGCUACUUUGUGUCAGC ...........................((((((((...((((((((((-(((((.(.....)...((((((...........)))))).)))))))))))...)))).)))))))).... ( -23.40) >consensus GAAAAUAAAUAAAAUAAAAUUAAAUUCGCAUAAAGAUGAGUAGAUAAU_AUUAAUUAAAAAGAGAUGUACUGGUUGGUGAAAAGUACAAUAAAUAUUGUUGAACGCUACUUUAUGU_CAA ...........................((((((((...(((.((((((.((((((..........((((((...........))))))))))))))))))....))).)))))))).... (-18.97 = -16.43 + -2.53)



| Location | 1,337,670 – 1,337,766 |
|---|---|
| Length | 96 |
| Sequences | 2 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 56.67 |
| Mean single sequence MFE | -26.41 |
| Consensus MFE | -17.10 |
| Energy contribution | -16.60 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1337670 96 + 2685015/160-280 --------UCUAUCAAGCGAU------AAUUUUUGCAGCUUAAAGAUAGAU----------UGUUUAUAACUAGGGUGGUACCGCGAAAACGUUCGUCCCUUUUUUUGAGGAUGAACGUU --------((((((((((...------..........))))...)))))).----------..............(((....)))...((((((((((((.......).))))))))))) ( -25.32) >sy_sa.0 1191327 117 + 2516575/160-280 AAUUAAAGUCAGUCGUUUGAUCUGUGUGAUACUUACAAUUAAGAUUGAAUUAAUUUGUUAACACUUAUAACUAGGGUGGUACCGCGA-AACGUUCGUCCCUUUAU--GAGGAUGGGCGUU ............(((((..((((.(((((...)))))....))))..).............(((((.......))))).....))))-((((((((((((.....--).))))))))))) ( -27.50) >consensus ________UCAAUCAAGCGAU______AAUACUUACAACUAAAAGAGAAAU__________CACUUAUAACUAGGGUGGUACCGCGA_AACGUUCGUCCCUUUAU__GAGGAUGAACGUU ...........................................................................(((....)))...((((((((((((.......).))))))))))) (-17.10 = -16.60 + -0.50)

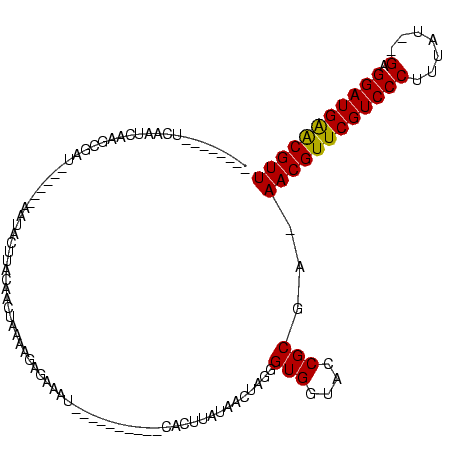
| Location | 1,337,670 – 1,337,775 |
|---|---|
| Length | 105 |
| Sequences | 2 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 64.41 |
| Mean single sequence MFE | -27.25 |
| Consensus MFE | -21.95 |
| Energy contribution | -21.70 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.81 |
| SVM decision value | 4.99 |
| SVM RNA-class probability | 0.999967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1337670 105 + 2685015/184-304 ---AAUUUUUGCAGCUUAAAGAUAGAU----------UGUUUAUAACUAGGGUGGUACCGCGAAAACGUUCGUCCCUUUUUUUGAGGAUGAACGUUUUUUUGUUUUUAGUUAAAAUAU ---.(((((((.....)))))))....----------(((((.(((((((((((....)))(((((((((((((((.......).)))))))))))))).....))))))))))))). ( -28.30) >sy_sa.0 1191327 115 + 2516575/184-304 UGUGAUACUUACAAUUAAGAUUGAAUUAAUUUGUUAACACUUAUAACUAGGGUGGUACCGCGA-AACGUUCGUCCCUUUAU--GAGGAUGGGCGUUUUUUAUUUUCUUUUUUAAUUAU ............((((((((..(((............(((((.......))))).......((-((((((((((((.....--).))))))))))))).....)))..)))))))).. ( -26.20) >consensus ___AAUACUUACAACUAAAAGAGAAAU__________CACUUAUAACUAGGGUGGUACCGCGA_AACGUUCGUCCCUUUAU__GAGGAUGAACGUUUUUUAGUUUCUAGUUAAAAUAU ................(((((((((..........................(((....)))(((((((((((((((.......).))))))))))))))))))))))).......... (-21.95 = -21.70 + -0.25)

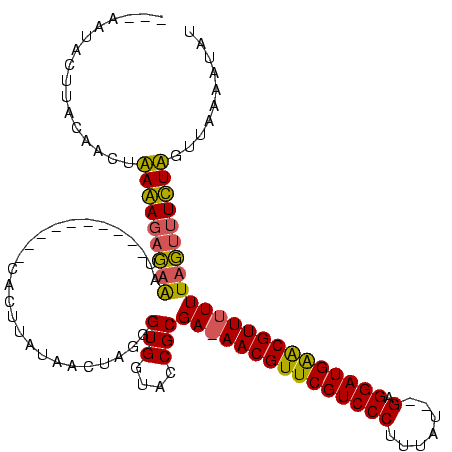
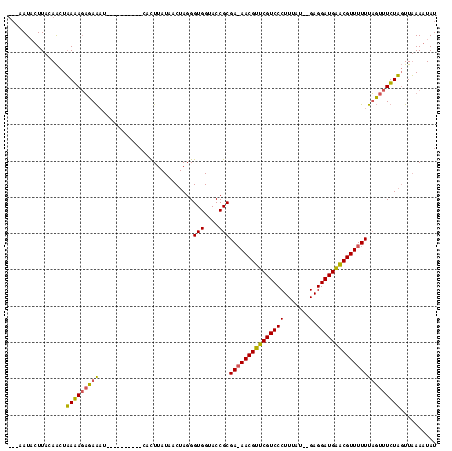
| Location | 1,337,670 – 1,337,775 |
|---|---|
| Length | 105 |
| Sequences | 2 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 64.41 |
| Mean single sequence MFE | -16.57 |
| Consensus MFE | -11.15 |
| Energy contribution | -11.65 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1337670 105 + 2685015/184-304 AUAUUUUAACUAAAAACAAAAAAACGUUCAUCCUCAAAAAAAGGGACGAACGUUUUCGCGGUACCACCCUAGUUAUAAACA----------AUCUAUCUUUAAGCUGCAAAAAUU--- ....................(((((((((.((((........)))).))))))))).(((((.......(((.........----------..))).......))))).......--- ( -19.44) >sy_sa.0 1191327 115 + 2516575/184-304 AUAAUUAAAAAAGAAAAUAAAAAACGCCCAUCCUC--AUAAAGGGACGAACGUU-UCGCGGUACCACCCUAGUUAUAAGUGUUAACAAAUUAAUUCAAUCUUAAUUGUAAGUAUCACA (((((((..............(((((..(.((((.--.....)))).)..))))-)...((.....)).)))))))..(((.((...((((((.......)))))).....)).))). ( -13.70) >consensus AUAAUUAAAAAAAAAAAAAAAAAACGCCCAUCCUC__AAAAAGGGACGAACGUU_UCGCGGUACCACCCUAGUUAUAAACA__________AACUAAAUCUAAACUGCAAAAAUC___ ..........((((......(((((((((.((((........)))).)))))))))...((.....))..............................))))................ (-11.15 = -11.65 + 0.50) # Strand winner: forward (1.00)

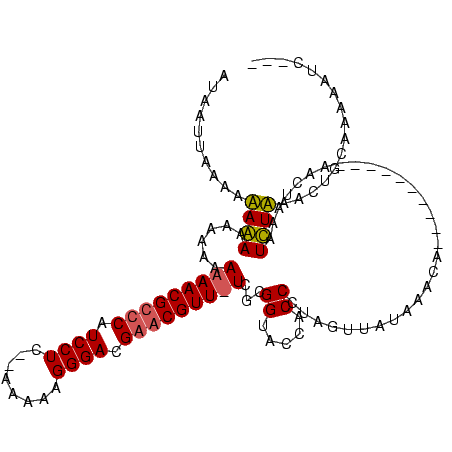
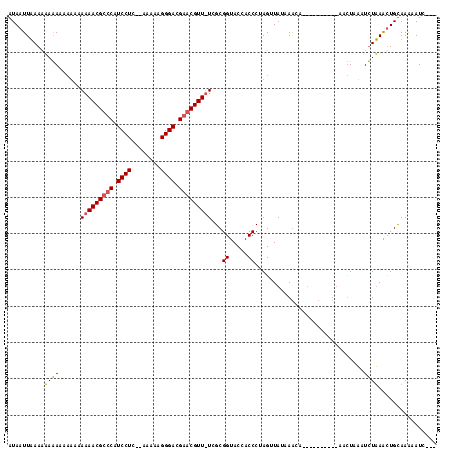
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:53:21 2006