| Sequence ID | sp_ag.0 |
|---|---|
| Location | 1,420,866 – 1,420,944 |
| Length | 78 |
| Max. P | 0.999697 |

| Location | 1,420,866 – 1,420,944 |
|---|---|
| Length | 78 |
| Sequences | 4 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 70.66 |
| Mean single sequence MFE | -12.78 |
| Consensus MFE | -11.40 |
| Energy contribution | -9.78 |
| Covariance contribution | -1.62 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.17 |
| SVM RNA-class probability | 0.998630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
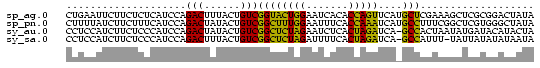
>sp_ag.0 1420866 78 - 2160267 CUGAAUUCUUCUCUCAUCCAGACUUUACUGUCGGUACUGGAAUCACACCAGUUCAUGCUCGAAAGCUCGCGGACUAUA ................((((((((........))).)))))........(((((.((..(....)..)).)))))... ( -14.30) >sp_pn.0 173890 78 + 2038615 CUUUUAUCUUCUUUCAUCCAGACUAUACUGUCGGCUUUGGAAUUUCACCAAAUCAUGCCUUUCGGCUCGUGGGCUAUA ....................(((......)))((((((((.......)))))....)))....((((....))))... ( -13.80) >sy_au.0 1565355 77 + 2809422 CCUCCAUCUUCUCCCAUCCAGACUAUACUGUCGGCUCUAGAAUCUCACUAGAUCA-GCCACUAAUAUGAUACAUACUA .....(((............(((......)))((((((((.......)))))...-)))........)))........ ( -11.80) >sy_sa.0 1211878 76 - 2516575 CCUCCAUCUUCUCCCAUCCAGACUUUACUGUCGGCUCUAGAUUUUCACUAGAUCA-GCCAUUU-UAUUAUAUAUAAUA ....................(((......)))((((((((.......)))))...-)))....-.............. ( -11.20) >consensus CCUCCAUCUUCUCCCAUCCAGACUAUACUGUCGGCUCUAGAAUCUCACCAGAUCA_GCCAUUAAGAUCAUAGACAAUA ....................(((......)))((((((((.......)))))....)))................... (-11.40 = -9.78 + -1.62)

| Location | 1,420,866 – 1,420,944 |
|---|---|
| Length | 78 |
| Sequences | 3 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 67.95 |
| Mean single sequence MFE | -13.10 |
| Consensus MFE | -11.20 |
| Energy contribution | -9.43 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.91 |
| SVM RNA-class probability | 0.999697 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
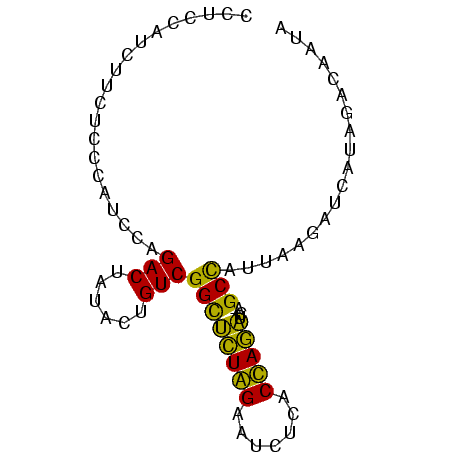
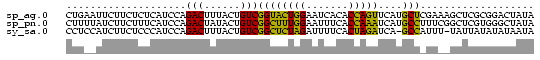
>sp_ag.0 1420866 78 - 2160267/0-78 CUGAAUUCUUCUCUCAUCCAGACUUUACUGUCGGUACUGGAAUCACACCAGUUCAUGCUCGAAAGCUCGCGGACUAUA ................((((((((........))).)))))........(((((.((..(....)..)).)))))... ( -14.30) >sp_pn.0 173890 78 + 2038615/0-78 CUUUUAUCUUCUUUCAUCCAGACUAUACUGUCGGCUUUGGAAUUUCACCAAAUCAUGCCUUUCGGCUCGUGGGCUAUA ....................(((......)))((((((((.......)))))....)))....((((....))))... ( -13.80) >sy_sa.0 1211878 76 - 2516575/0-78 CCUCCAUCUUCUCCCAUCCAGACUUUACUGUCGGCUCUAGAUUUUCACUAGAUCA-GCCAUUU-UAUUAUAUAUAAUA ....................(((......)))((((((((.......)))))...-)))....-.............. ( -11.20) >consensus CUUAAAUCUUCUCUCAUCCAGACUUUACUGUCGGCUCUGGAAUUUCACCAGAUCAUGCCAUUA_GCUCGUGGACUAUA ....................(((......)))((((((((.......)))))....)))................... (-11.20 = -9.43 + -1.77)
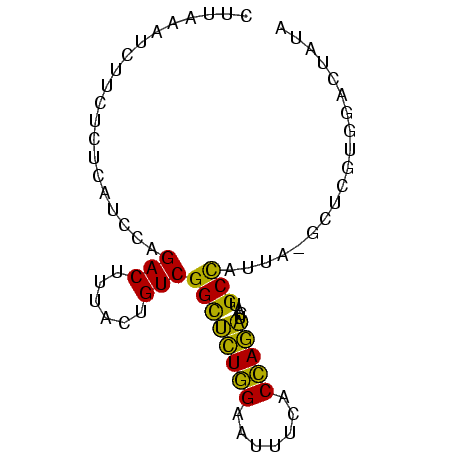
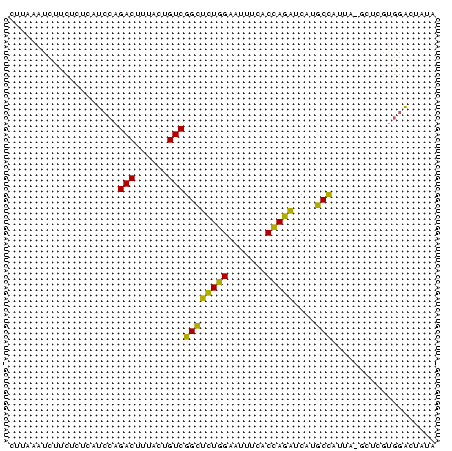
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:51:06 2006