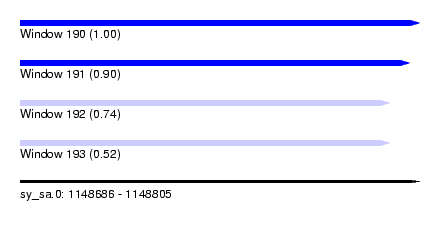
| Sequence ID | sy_sa.0 |
|---|---|
| Location | 1,148,686 – 1,148,805 |
| Length | 119 |
| Max. P | 0.999820 |

| Location | 1,148,686 – 1,148,805 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 64.64 |
| Mean single sequence MFE | -20.40 |
| Consensus MFE | -14.64 |
| Energy contribution | -12.20 |
| Covariance contribution | -2.44 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.72 |
| SVM decision value | 4.16 |
| SVM RNA-class probability | 0.999820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 1148686 119 + 2516575/120-240 CAAGCACUUAAUUACAUGAUUAACGCUCAAGCACGCUUUAACCUAUUAAACAGAUAUCUUCAUAUGAUUAGUGUUCAGUUAAAGUUCAAUGUGGGCUACCUUCA-AUAAGUUAAAUUGUA ...(((.((((((...(((.....(((((.....((((((((......((((...(((.......)))...))))..))))))))......))))).....)))-...))))))..))). ( -18.70) >sy_ha.0 1298497 99 + 2685015/120-240 CUUGCACUUAAUU---UGAUUAACGCUCAAACACGCUUUUAUCUACUAA---------UUAAUG--------GUUCAAUAAAAGUUCAA-GUGGGCUACCUUCAGAUUGAAUGUUAUACA ...(((.((((((---(((.....(((((.....((((((((..((((.---------....))--------))...))))))))....-.))))).....))))))))).)))...... ( -23.90) >sy_au.0 1066761 104 - 2809422/120-240 CAAGCACUCUAGAUUAUGAUUAACGCUCAAACACGUCUUAGCCUACUA----------UUAAUC------ACGUUCAGCUAAGAUACUCUGUGGGCUACCUUCAGUAAAAAUCAUUUACA ................(((.....(((((.....((((((((..((..----------......------..))...))))))))......))))).....)))(((((.....))))). ( -18.60) >consensus CAAGCACUUAAUU__AUGAUUAACGCUCAAACACGCUUUAACCUACUAA_________UUAAUA________GUUCAGUUAAAGUUCAAUGUGGGCUACCUUCAGAUAAAAUAAUUUACA ................(((.....(((((.....((((((((..((..........................))...))))))))......))))).....)))................ (-14.64 = -12.20 + -2.44) # Strand winner: reverse (0.98)

| Location | 1,148,686 – 1,148,802 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 61.34 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -10.58 |
| Energy contribution | -10.27 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.45 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.34 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 1148686 116 + 2516575/200-320 UAAAGCGUGCUUGAGCGUUAAUCAUGUAAUUAAGUGCUUGAA-AU-UAGUUUGUAAUUUCUUGAAUUUAGGUGGUACCACG--GAAUAUCCGUCCUAUUUUGAUAGGGCGGAUAUUUUUU ......(..(((((((((.....))))..)))))..)..(((-((-((.....)))))))..........(((....)))(--(((((((((((((((....)))))))))))))))).. ( -35.20) >sy_ha.0 1298497 110 + 2685015/200-320 AAAAGCGUGUUUGAGCGUUAAUCA---AAUUAAGUGCAAGUAUAU-UAAUUUAUA----CUUGAAUUUAGGUGGUACCACG--GAACAUCCGUCCUAUAUUAUUUAGGAGGAUGUUUUUU ...(((((......))))).....---..((((((.((((((((.-.....))))----)))).))))))(((....)))(--((((((((.(((((.......)))))))))))))).. ( -34.50) >sy_au.0 1066761 120 - 2809422/200-320 UAAGACGUGUUUGAGCGUUAAUCAUAAUCUAGAGUGCUUGAAGAGAUGAAGCAUUUCAAAUUUCACGAUGAUAGUAUUGGUAUGAUUAUUCCAAUUAUGUUAAAAGAGUGAAUUUGAUAU ...(((((......))))).............(((((((.(.....).)))))))(((((((.(((..(((((..(((((..........)))))..))))).....))))))))))... ( -22.70) >consensus UAAAGCGUGUUUGAGCGUUAAUCAU__AAUUAAGUGCUUGAA_AU_UAAUUUAUA____CUUGAAUUUAGGUGGUACCACG__GAAUAUCCGUCCUAUAUUAAUAGGGAGGAUAUUUUUU ...(((((......)))))...................................................(((....)))...((((((((.((((.........))))))))))))... (-10.58 = -10.27 + -0.32)

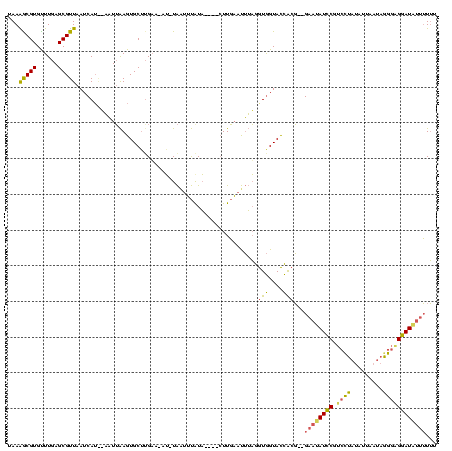
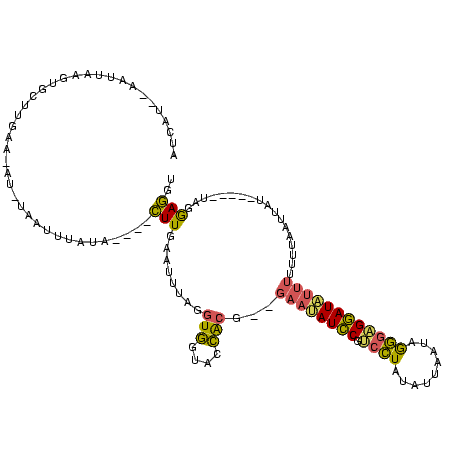
| Location | 1,148,686 – 1,148,796 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 54.55 |
| Mean single sequence MFE | -29.43 |
| Consensus MFE | -6.57 |
| Energy contribution | -6.03 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.50 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.22 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 1148686 110 + 2516575/220-340 AUCAUGUAAUUAAGUGCUUGAA-AU-UAGUUUGUAAUUUCUUGAAUUUAGGUGGUACCACG--GAAUAUCCGUCCUAUUUUGAUAGGGCGGAUAUUUUUU-AUUUUU-----AAGGAGGA .(((.(((......))).))).-..-..........(((((((((..((((((....)))(--(((((((((((((((....))))))))))))))))))-)..)))-----)))))).. ( -34.00) >sy_ha.0 1298497 105 + 2685015/220-340 AUCA---AAUUAAGUGCAAGUAUAU-UAAUUUAUA----CUUGAAUUUAGGUGGUACCACG--GAACAUCCGUCCUAUAUUAUUUAGGAGGAUGUUUUUUUAAUUAU-----UAGGAGGU ....---..((((((.((((((((.-.....))))----)))).))))))(((....)))(--((((((((.(((((.......)))))))))))))).........-----........ ( -30.60) >sy_au.0 1066761 120 - 2809422/220-340 AUCAUAAUCUAGAGUGCUUGAAGAGAUGAAGCAUUUCAAAUUUCACGAUGAUAGUAUUGGUAUGAUUAUUCCAAUUAUGUUAAAAGAGUGAAUUUGAUAUUAAUCAUCCUUCUUGAAUAU ................(..((((.(((((......(((((((.(((..(((((..(((((..........)))))..))))).....))))))))))......)))))))))..)..... ( -23.70) >consensus AUCAU__AAUUAAGUGCUUGAA_AU_UAAUUUAUA____CUUGAAUUUAGGUGGUACCACG__GAAUAUCCGUCCUAUAUUAAUAGGGAGGAUAUUUUUUUAAUUAU_____UAGGAGGU .......................................(((........(((....)))...((((((((.((((.........))))))))))))..................))).. ( -6.57 = -6.03 + -0.54)

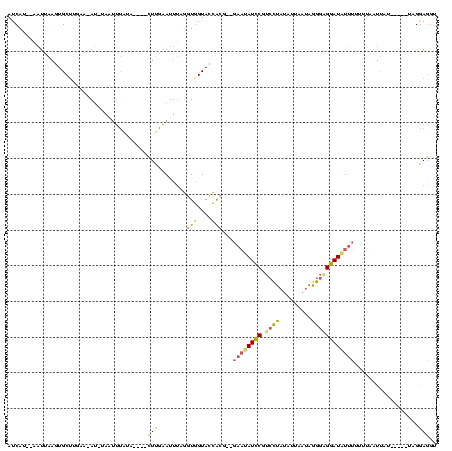
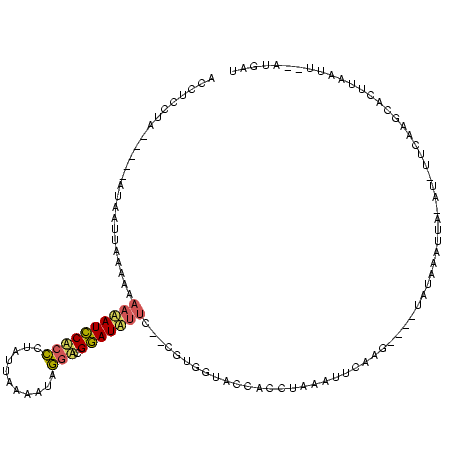
| Location | 1,148,686 – 1,148,796 |
|---|---|
| Length | 110 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 54.55 |
| Mean single sequence MFE | -21.63 |
| Consensus MFE | -3.23 |
| Energy contribution | -3.57 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.40 |
| Mean z-score | -3.75 |
| Structure conservation index | 0.15 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_sa.0 1148686 110 + 2516575/220-340 UCCUCCUU-----AAAAAU-AAAAAAUAUCCGCCCUAUCAAAAUAGGACGGAUAUUC--CGUGGUACCACCUAAAUUCAAGAAAUUACAAACUA-AU-UUCAAGCACUUAAUUACAUGAU ........-----......-....((((((((.(((((....))))).)))))))).--.(((....)))..........(((((((.....))-))-)))................... ( -21.10) >sy_ha.0 1298497 105 + 2685015/220-340 ACCUCCUA-----AUAAUUAAAAAAACAUCCUCCUAAAUAAUAUAGGACGGAUGUUC--CGUGGUACCACCUAAAUUCAAG----UAUAAAUUA-AUAUACUUGCACUUAAUU---UGAU ........-----...((((((..((((((((((((.......))))).))))))).--.(((....))).......((((----((((.....-.)))))))).......))---)))) ( -23.40) >sy_au.0 1066761 120 - 2809422/220-340 AUAUUCAAGAAGGAUGAUUAAUAUCAAAUUCACUCUUUUAACAUAAUUGGAAUAAUCAUACCAAUACUAUCAUCGUGAAAUUUGAAAUGCUUCAUCUCUUCAAGCACUCUAGAUUAUGAU ........(((((((((...((.((((((((((............(((((.((....)).))))).........))).))))))).))...))))).))))................... ( -20.40) >consensus ACCUCCUA_____AUAAUUAAAAAAAAAUCCACCCUAUUAAAAUAGGACGGAUAUUC__CGUGGUACCACCUAAAUUCAAG____UAUAAAUUA_AU_UUCAAGCACUUAAUU__AUGAU ........................((((((((((...........))).)))))))................................................................ ( -3.23 = -3.57 + 0.34) # Strand winner: forward (0.91)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:53:16 2006