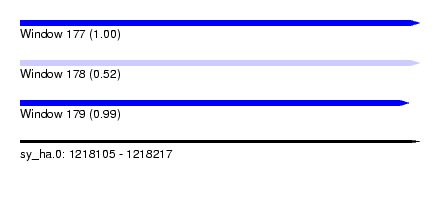
| Sequence ID | sy_ha.0 |
|---|---|
| Location | 1,218,105 – 1,218,217 |
| Length | 112 |
| Max. P | 0.999291 |

| Location | 1,218,105 – 1,218,217 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 68.18 |
| Mean single sequence MFE | -35.63 |
| Consensus MFE | -16.18 |
| Energy contribution | -17.30 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.22 |
| Mean z-score | -4.68 |
| Structure conservation index | 0.45 |
| SVM decision value | 3.49 |
| SVM RNA-class probability | 0.999291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1218105 112 + 2685015/200-320 -CGUUU--AUUAAUGAACGUAUCAUUGAUAGAGUGAAUAGCAAUAUGC-----UAUUUAAGUAUGGUGGUAACGUGCUCAAGCGCCCUUACAUUUAACGAUGUAUGGGUGCUUUUUAUUU -..(((--((((((((.....))))))))))).((((((((.....))-----))))))((((((.......)))))).((((((((.((((((....)))))).))))))))....... ( -39.10) >sy_au.0 989159 116 - 2809422/200-320 -CGUUU--AUUAAUGAACGUAUCAUUAAUAGAGUGAACAUUAUCUUAUGAUUAUGUUUAAGUUUGGUGGUAACGUGCAAUCGCGCCCUUACAUAUU-UGUUGUAAGGGUUUUUUUUACGU -(((((--(....))))))(((((((((.....(((((((.(((....))).)))))))...)))))))))(((((.((....(((((((((....-...)))))))))...)).))))) ( -33.80) >sy_sa.0 1070124 107 + 2516575/200-320 ACGUUUUAGUUAAUGAACGUAUCAUUAAUAAAGU------UAUUUGAC-------UUAAAGUAUGGUGGUACCGUGCGCAUGCGCCCUUACAUUAAUUUGUAAGGGCGUUUUUCUAUUUU ........((((((((.....)))))))).((((------(....)))-------))...((((((.....))))))....(((((((((((......)))))))))))........... ( -34.00) >consensus _CGUUU__AUUAAUGAACGUAUCAUUAAUAGAGUGAA_A_UAUUUUAC_____U_UUUAAGUAUGGUGGUAACGUGCACAAGCGCCCUUACAUUUA_UGAUGUAAGGGUUUUUUUUAUUU ........((((((((.....))))))))...............................(((((.......)))))......((((((((((......))))))))))........... (-16.18 = -17.30 + 1.12)

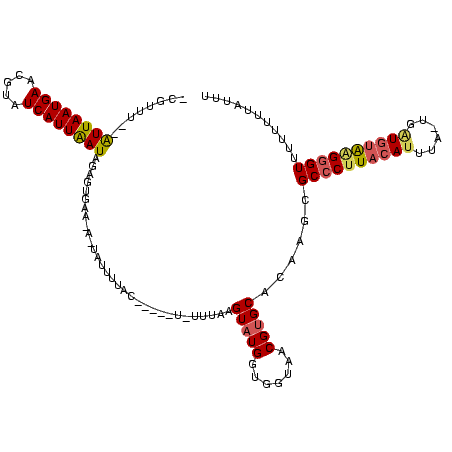
| Location | 1,218,105 – 1,218,217 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.18 |
| Mean single sequence MFE | -24.50 |
| Consensus MFE | -7.63 |
| Energy contribution | -8.30 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.86 |
| Structure conservation index | 0.31 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.521987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1218105 112 + 2685015/200-320 AAAUAAAAAGCACCCAUACAUCGUUAAAUGUAAGGGCGCUUGAGCACGUUACCACCAUACUUAAAUA-----GCAUAUUGCUAUUCACUCUAUCAAUGAUACGUUCAUUAAU--AAACG- .......((((.(((.(((((......))))).))).)))).....((((.............((((-----((.....)))))).....(((.(((((.....))))).))--)))))- ( -24.20) >sy_au.0 989159 116 - 2809422/200-320 ACGUAAAAAAAACCCUUACAACA-AAUAUGUAAGGGCGCGAUUGCACGUUACCACCAAACUUAAACAUAAUCAUAAGAUAAUGUUCACUCUAUUAAUGAUACGUUCAUUAAU--AAACG- ..((((......((((((((...-....)))))))).....)))).((((.............(((((.(((....))).))))).....(((((((((.....))))))))--)))))- ( -25.00) >sy_sa.0 1070124 107 + 2516575/200-320 AAAAUAGAAAAACGCCCUUACAAAUUAAUGUAAGGGCGCAUGCGCACGGUACCACCAUACUUUAA-------GUCAAAUA------ACUUUAUUAAUGAUACGUUCAUUAACUAAAACGU ............((((((((((......))))))))))...(((...((.....)).........-------........------..(((((((((((.....))))))).)))).))) ( -24.30) >consensus AAAUAAAAAAAACCCAUACAACA_UAAAUGUAAGGGCGCAUGAGCACGUUACCACCAUACUUAAA_A_____GUAAAAUA_U_UUCACUCUAUUAAUGAUACGUUCAUUAAU__AAACG_ .............................((((.(((......)).).))))........................................(((((((.....)))))))......... ( -7.63 = -8.30 + 0.67) # Strand winner: forward (1.00)

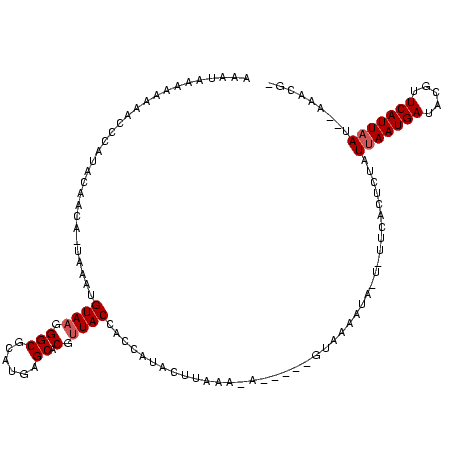
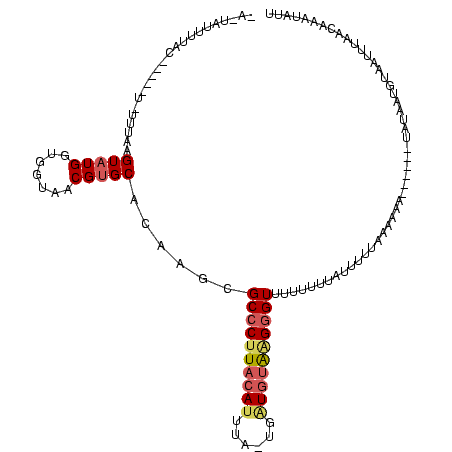
| Location | 1,218,105 – 1,218,214 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 58.00 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -10.81 |
| Energy contribution | -12.37 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.20 |
| Mean z-score | -4.21 |
| Structure conservation index | 0.34 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1218105 109 + 2685015/237-357 UAGCAAUAUGC-----UAUUUAAGUAUGGUGGUAACGUGCUCAAGCGCCCUUACAUUUAACGAUGUAUGGGUGCUUUUUAUUUUUCAUAAA------UAGAGUAUAGAUCACUAAAUAUU ((((.....))-----))........(((((((...((((((((((((((.((((((....)))))).))))))))..(((((.....)))------))))))))..)))))))...... ( -36.20) >sy_au.0 989159 114 - 2809422/237-357 CAUUAUCUUAUGAUUAUGUUUAAGUUUGGUGGUAACGUGCAAUCGCGCCCUUACAUAUU-UGUUGUAAGGGUUUUUUUUACGUUCAAAAAAC-----UUUAAAGUAUUUUAACAAGUAUU (((.(((....))).)))...((((((..(((..(((((.((....(((((((((....-...)))))))))...)).))))))))..))))-----))...(((((((....))))))) ( -25.00) >sy_sa.0 1070124 110 + 2516575/237-357 ---UAUUUGAC-------UUAAAGUAUGGUGGUACCGUGCGCAUGCGCCCUUACAUUAAUUUGUAAGGGCGUUUUUCUAUUUUUUUAGAAAUCUAUAUAUGUUGCAAUUUAACACAUACU ---........-------....((((((.((......((((((((((((((((((......))))))))))).((((((......)))))).......))).))))......)))))))) ( -35.10) >consensus _A_UAUUUUAC_____U_UUUAAGUAUGGUGGUAACGUGCACAAGCGCCCUUACAUUUA_UGAUGUAAGGGUUUUUUUUAUUUUUAAAAAA______UAUAAUGUAAUUUAACAAAUAUU .......................(((((.......)))))......((((((((((......))))))))))................................................ (-10.81 = -12.37 + 1.56)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:53:05 2006