
| Sequence ID | sy_sa.0 |
|---|---|
| Location | 1,046,773 – 1,046,859 |
| Length | 86 |
| Max. P | 0.999685 |

| Location | 1,046,773 – 1,046,859 |
|---|---|
| Length | 86 |
| Sequences | 3 |
| Columns | 86 |
| Reading direction | forward |
| Mean pairwise identity | 78.68 |
| Mean single sequence MFE | -22.30 |
| Consensus MFE | -17.44 |
| Energy contribution | -18.67 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.78 |
| SVM decision value | 3.72 |
| SVM RNA-class probability | 0.999559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
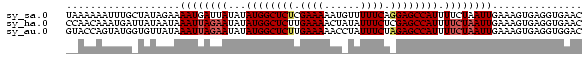
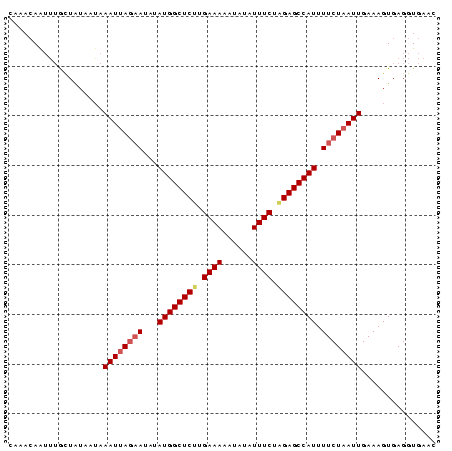
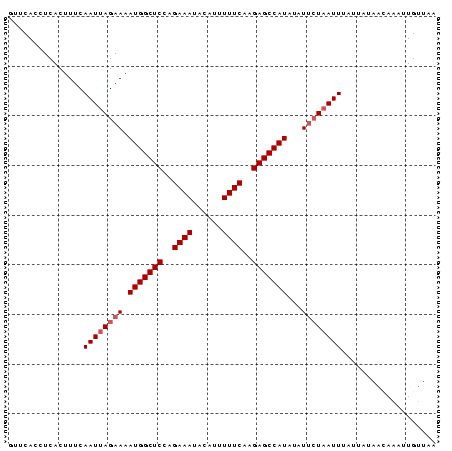
>sy_sa.0 1046773 86 + 2516575 UAAAAAAUUUGCUAUAGAAAAUGAUUAUAUAUGGCUCUCGAAAAAUGUUUUUCAGGAGCCAUUUUCUAAUUGAAAGUGAGGUGAAC .......((..((...................(((((..(((((....)))))..)))))((((((.....))))))..))..)). ( -20.00) >sy_ha.0 1194877 86 + 2685015 CCAACAAAUGAUUAUAAUAAAUUAGAAUAUAUGGCUCUUGAAAACUAUAUUUCUCGAGCCAUUUUCUAAUUGAAAGUGAGGUGAAC ....((.....((((....((((((((...(((((((..((((......))))..))))))).))))))))....))))..))... ( -21.80) >sy_au.0 962051 86 - 2809422 GUACCAGUAUGGUGUUAUAAAUUAGAAUAUAUGGCUCUUGAAAAACCUAUUUCUAGAGCCAUUUUCUAAUUGAAAGUGAGGUGGAC .((((.(((......))).((((((((...((((((((.((((......)))).)))))))).))))))))........))))... ( -25.10) >consensus CAAACAAUUUGCUAUAAUAAAUUAGAAUAUAUGGCUCUUGAAAAAUAUAUUUCUAGAGCCAUUUUCUAAUUGAAAGUGAGGUGAAC ...................((((((((...((((((((.((((......)))).)))))))).))))))))............... (-17.44 = -18.67 + 1.22)

| Location | 1,046,773 – 1,046,859 |
|---|---|
| Length | 86 |
| Sequences | 3 |
| Columns | 86 |
| Reading direction | reverse |
| Mean pairwise identity | 78.68 |
| Mean single sequence MFE | -19.60 |
| Consensus MFE | -15.13 |
| Energy contribution | -16.13 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.70 |
| Structure conservation index | 0.77 |
| SVM decision value | 3.89 |
| SVM RNA-class probability | 0.999685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
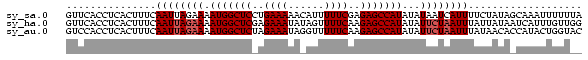
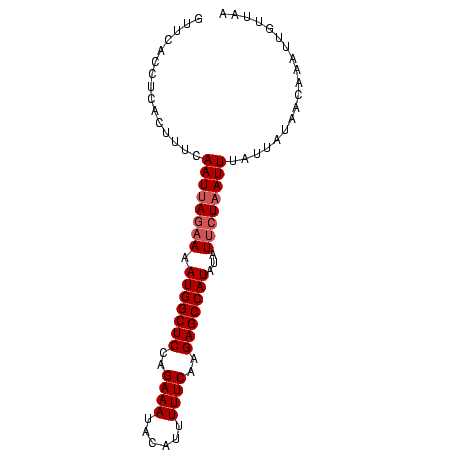
>sy_sa.0 1046773 86 + 2516575 GUUCACCUCACUUUCAAUUAGAAAAUGGCUCCUGAAAAACAUUUUUCGAGAGCCAUAUAUAAUCAUUUUCUAUAGCAAAUUUUUUA ..................(((((((((((((.((((((....)))))).)))))))..........)))))).............. ( -16.50) >sy_ha.0 1194877 86 + 2685015 GUUCACCUCACUUUCAAUUAGAAAAUGGCUCGAGAAAUAUAGUUUUCAAGAGCCAUAUAUUCUAAUUUAUUAUAAUCAUUUGUUGG ...............((((((((.(((((((..((((......))))..)))))))...))))))))................... ( -19.40) >sy_au.0 962051 86 - 2809422 GUCCACCUCACUUUCAAUUAGAAAAUGGCUCUAGAAAUAGGUUUUUCAAGAGCCAUAUAUUCUAAUUUAUAACACCAUACUGGUAC ...............((((((((.((((((((.((((......)))).))))))))...))))))))......(((.....))).. ( -22.90) >consensus GUUCACCUCACUUUCAAUUAGAAAAUGGCUCCAGAAAUACAUUUUUCAAGAGCCAUAUAUUCUAAUUUAUUAUAACAAAUUGUUAA ...............((((((((.(((((((..((((......))))..)))))))...))))))))................... (-15.13 = -16.13 + 1.00) # Strand winner: forward (0.99)
| Location | 1,046,773 – 1,046,859 |
|---|---|
| Length | 86 |
| Sequences | 3 |
| Columns | 86 |
| Reading direction | forward |
| Mean pairwise identity | 78.68 |
| Mean single sequence MFE | -22.30 |
| Consensus MFE | -17.44 |
| Energy contribution | -18.67 |
| Covariance contribution | 1.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.78 |
| SVM decision value | 3.72 |
| SVM RNA-class probability | 0.999559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
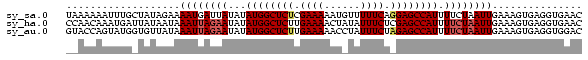
>sy_sa.0 1046773 86 + 2516575/0-86 UAAAAAAUUUGCUAUAGAAAAUGAUUAUAUAUGGCUCUCGAAAAAUGUUUUUCAGGAGCCAUUUUCUAAUUGAAAGUGAGGUGAAC .......((..((...................(((((..(((((....)))))..)))))((((((.....))))))..))..)). ( -20.00) >sy_ha.0 1194877 86 + 2685015/0-86 CCAACAAAUGAUUAUAAUAAAUUAGAAUAUAUGGCUCUUGAAAACUAUAUUUCUCGAGCCAUUUUCUAAUUGAAAGUGAGGUGAAC ....((.....((((....((((((((...(((((((..((((......))))..))))))).))))))))....))))..))... ( -21.80) >sy_au.0 962051 86 - 2809422/0-86 GUACCAGUAUGGUGUUAUAAAUUAGAAUAUAUGGCUCUUGAAAAACCUAUUUCUAGAGCCAUUUUCUAAUUGAAAGUGAGGUGGAC .((((.(((......))).((((((((...((((((((.((((......)))).)))))))).))))))))........))))... ( -25.10) >consensus CAAACAAUUUGCUAUAAUAAAUUAGAAUAUAUGGCUCUUGAAAAAUAUAUUUCUAGAGCCAUUUUCUAAUUGAAAGUGAGGUGAAC ...................((((((((...((((((((.((((......)))).)))))))).))))))))............... (-17.44 = -18.67 + 1.22)


| Location | 1,046,773 – 1,046,859 |
|---|---|
| Length | 86 |
| Sequences | 3 |
| Columns | 86 |
| Reading direction | reverse |
| Mean pairwise identity | 78.68 |
| Mean single sequence MFE | -19.60 |
| Consensus MFE | -15.13 |
| Energy contribution | -16.13 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.70 |
| Structure conservation index | 0.77 |
| SVM decision value | 3.89 |
| SVM RNA-class probability | 0.999685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
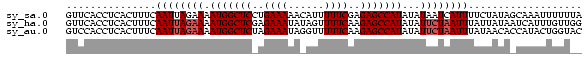
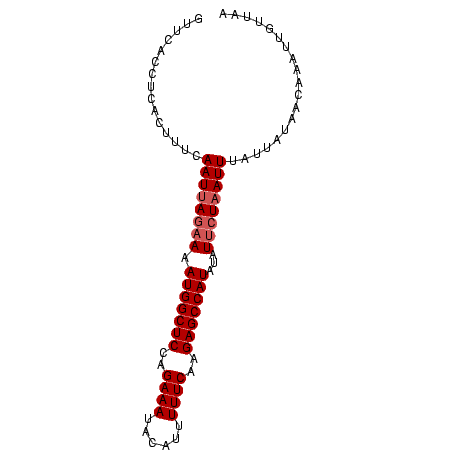
>sy_sa.0 1046773 86 + 2516575/0-86 GUUCACCUCACUUUCAAUUAGAAAAUGGCUCCUGAAAAACAUUUUUCGAGAGCCAUAUAUAAUCAUUUUCUAUAGCAAAUUUUUUA ..................(((((((((((((.((((((....)))))).)))))))..........)))))).............. ( -16.50) >sy_ha.0 1194877 86 + 2685015/0-86 GUUCACCUCACUUUCAAUUAGAAAAUGGCUCGAGAAAUAUAGUUUUCAAGAGCCAUAUAUUCUAAUUUAUUAUAAUCAUUUGUUGG ...............((((((((.(((((((..((((......))))..)))))))...))))))))................... ( -19.40) >sy_au.0 962051 86 - 2809422/0-86 GUCCACCUCACUUUCAAUUAGAAAAUGGCUCUAGAAAUAGGUUUUUCAAGAGCCAUAUAUUCUAAUUUAUAACACCAUACUGGUAC ...............((((((((.((((((((.((((......)))).))))))))...))))))))......(((.....))).. ( -22.90) >consensus GUUCACCUCACUUUCAAUUAGAAAAUGGCUCCAGAAAUACAUUUUUCAAGAGCCAUAUAUUCUAAUUUAUUAUAACAAAUUGUUAA ...............((((((((.(((((((..((((......))))..)))))))...))))))))................... (-15.13 = -16.13 + 1.00) # Strand winner: forward (0.99)
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:53:02 2006