| Sequence ID | sy_ha.0 |
|---|---|
| Location | 1,160,034 – 1,160,154 |
| Length | 120 |
| Max. P | 0.997901 |

| Location | 1,160,034 – 1,160,154 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.37 |
| Mean single sequence MFE | -23.68 |
| Consensus MFE | -18.20 |
| Energy contribution | -18.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.800785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1160034 120 + 2685015/120-240 AAGACGUCUCAUUCUUUCUCCCAUCCAGACUUUCACUGUCGGCUCUAGAAUCUCACUAGAUCAGCCAUCAAAGUUAUUACCUUUCACUUUGACAGGUCGCAGGCUUAUAUUACUGCCGGU ..(((((((.................)))).....(((((((((((((.......)))))...)))...(((((...........)))))))))))))((((..........)))).... ( -25.33) >sy_au.0 933115 114 - 2809422/120-240 GAUAAUUAACAUUCUUUCUCCCAUCCAGACUUUAACUGUCGGCUCUAGAAUCUCACUAGAUCAGCCACUAA------UAUGAAACAUAUUAGCAGGUCGCAGGCUUUAUUUACUGCCGGU ....................((.....(((((........((((((((.......)))))...))).((((------((((...)))))))).)))))((((..........)))).)). ( -23.90) >sy_sa.0 1025602 112 + 2516575/120-240 GUUAUACUUCAUUCUUUCUCCCAUCCAGACUUU-ACUGUCGGCUCUAGAAUCUCACUAGAUCAGCCAC-------AUUUAUACACAUAAAUGCAGGUCGCAGGCUUAAUUUACUGCCGGU ....................((.....(((((.-......((((((((.......)))))...))).(-------((((((....))))))).)))))((((..........)))).)). ( -21.80) >consensus GAUAAAUAUCAUUCUUUCUCCCAUCCAGACUUU_ACUGUCGGCUCUAGAAUCUCACUAGAUCAGCCAC_AA____AUUAACAAACAUAUUAGCAGGUCGCAGGCUUAAUUUACUGCCGGU ....................((..((.(((.......)))((((((((.......)))))...)))............................))..((((..........)))).)). (-18.20 = -18.20 + -0.00) # Strand winner: forward (0.76)

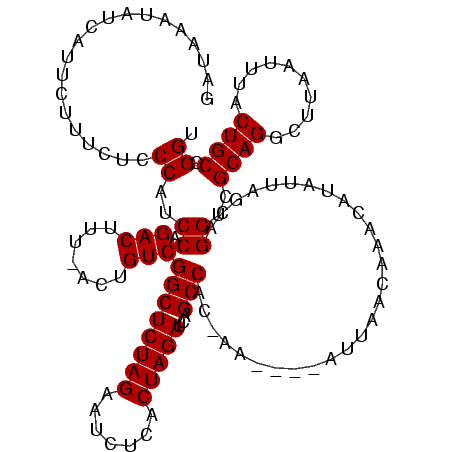
| Location | 1,160,034 – 1,160,149 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.66 |
| Mean single sequence MFE | -17.73 |
| Consensus MFE | -10.70 |
| Energy contribution | -10.70 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.96 |
| SVM RNA-class probability | 0.997901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 1160034 115 + 2685015/160-280 UGU---GUAUGCUAAAUAACCGUC--AUAAUUAUCAAAUAAAGACGUCUCAUUCUUUCUCCCAUCCAGACUUUCACUGUCGGCUCUAGAAUCUCACUAGAUCAGCCAUCAAAGUUAUUAC ...---........((((((((((--................)))).....................(((.......)))((((((((.......)))))...)))......)))))).. ( -15.99) >sy_au.0 933115 106 - 2809422/160-280 CAU---UUUUAC-AAAUACGCUAC--AUUAUCUUU--GUCGAUAAUUAACAUUCUUUCUCCCAUCCAGACUUUAACUGUCGGCUCUAGAAUCUCACUAGAUCAGCCACUAA------UAU ...---......-......(((..--.........--(((((((.((((...(((...........)))..)))).)))))))(((((.......)))))..)))......------... ( -13.90) >sy_sa.0 1025602 112 + 2516575/160-280 UUUAUGGUAUGCCAAAUAAACACUUAAAAAGUGUUGUUUUGUUAUACUUCAUUCUUUCUCCCAUCCAGACUUU-ACUGUCGGCUCUAGAAUCUCACUAGAUCAGCCAC-------AUUUA .....((((((.((((..((((((.....))))))..)))).))))))...................(((...-...)))((((((((.......)))))...)))..-------..... ( -23.30) >consensus UAU___GUAUGC_AAAUAAACAAC__AUAAUUAUU__AUAGAUAAAUAUCAUUCUUUCUCCCAUCCAGACUUU_ACUGUCGGCUCUAGAAUCUCACUAGAUCAGCCAC_AA____AUUAA ...................................................................(((.......)))((((((((.......)))))...))).............. (-10.70 = -10.70 + 0.00) # Strand winner: forward (0.69)

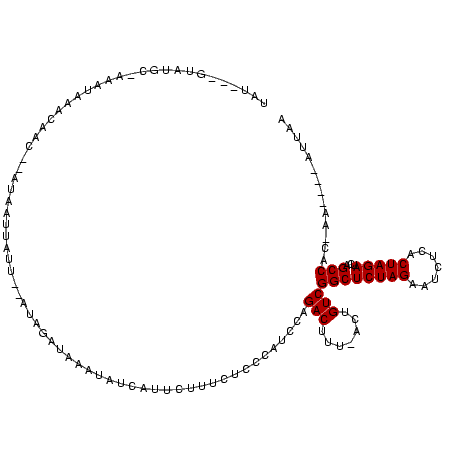
| Location | 1,160,034 – 1,160,135 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 50.88 |
| Mean single sequence MFE | -23.03 |
| Consensus MFE | -7.97 |
| Energy contribution | -7.76 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.70 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.35 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938298 |
| Prediction | RNA |
| WARNING | Mean pairwise identity too low. |
Download alignment: ClustalW | MAF
>sy_ha.0 1160034 101 + 2685015/269-389 CAUAC---ACAUUUCUAAUGU---UGUAAGCUUAUUUGA------AUAUACAUUUGGUCUCCCUAUGCCUUU-AAGCACUUUAUUGGUAUAGGGAGUUUUAAUUUU------GAGGUGAU .....---.((((((.(((((---.(((..(......).------.))))))))....(((((((((((..(-(((...))))..)))))))))))..........------)))))).. ( -26.70) >sy_au.0 933115 91 - 2809422/269-389 UAAAA---AUGUGUACAA-------AUAGGCUUAUUUAA------CGAUAAAUUUU-UCUCCUUGCAUCUUA-AUUCA------UGAUGUGAGGAUUUUUGUUUAUA-----GAGGUGAU .....---..........-------......((((((..------..((((((...-..((((..((((...-.....------.))))..)))).....)))))).-----.)))))). ( -18.10) >sy_sa.0 1025602 120 + 2516575/269-389 CAUACCAUAAAUUUAAAAUUUAUGUAUAAGUGUGUCUAAUUUUUGCUAAUAAUGUUAUAUCAAUAGAUCCCACACCUGAAAUGUUCAUUUCAGGUGUUUUUUUUAUUAUUUAGAGGUGAU ((((((((((((.....))))))).....)))))(((((.....((.......))......((((((....(((((((((((....)))))))))))....))))))..)))))...... ( -24.30) >consensus CAUAC___AAAUUUAAAAU_U___UAUAAGCUUAUUUAA______CUAUAAAUUUU_UCUCCAUACAUCUUA_AACCA___U_UUGAUAUAAGGAGUUUUAUUUAU______GAGGUGAU ...........................................................((((((((((................))))))))))......................... ( -7.97 = -7.76 + -0.21)



Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:52:58 2006