
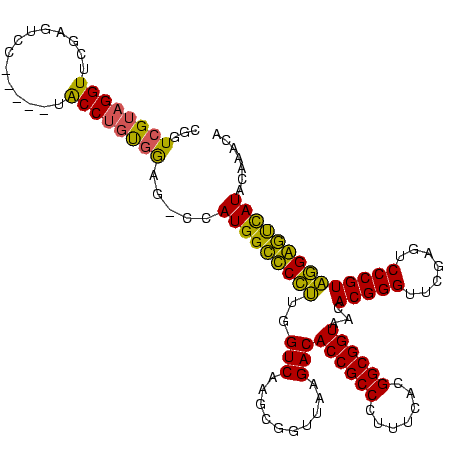
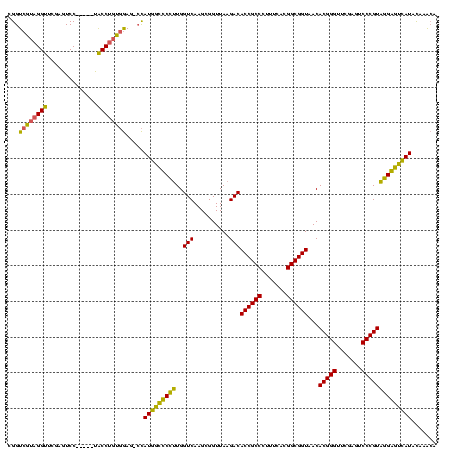
| Sequence ID | sy_au.0 |
|---|---|
| Location | 1,940,108 – 1,940,228 |
| Length | 120 |
| Max. P | 0.999980 |

| Location | 1,940,108 – 1,940,222 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.90 |
| Mean single sequence MFE | -43.05 |
| Consensus MFE | -38.18 |
| Energy contribution | -36.37 |
| Covariance contribution | -1.81 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.534129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1940108 114 + 2809422/0-120 CGGUCGUAGGUUCGAGUCC-----UACCUGUGGAG-CCAUGGCUCCUUGGUCAAGCGGUUAAGACACCGCCCUUUCACGGCGGUAACACGGGUUCGAGUCCCGUAGGAGUCAUACAAGCA .(..((((((.......))-----)))....((((-(....)))))..)..)..((...((.(((((((((.......))))))...(((((.......)))))....))).))...)). ( -41.70) >sy_sa.0 1512280 114 - 2516575/0-120 CGGUCGUAGGUUCGAGUCC-----UACCUGUGGAG-CCAUGGCCCCGUGGUCAAGCGGUUAAGACACCGCCCUUUCACGGCGGUAACACGGGUUCGAGUCCCGUCGGGGUCAUAUAAACA .((((((((((........-----.)))))))..)-))((((((((((((....(((((......)))))....))))(((((..((.((....)).)).)))))))))))))....... ( -42.30) >sy_ha.0 1543851 114 - 2685015/0-120 CGGUCGUAGGUUCGAAUCC-----UACCUGCGGAG-CCAUGGCUCCUUGGUCAAGCGGUUAAGACACCGCCCUUUCACGGCGGUAACACGGGUUCGAGUCCCGUAGGAGUCAUACAAACA .((((((((((........-----.)))))))..)-))(((((((((..(((..........)))((((((.......))))))...(((((.......))))))))))))))....... ( -43.50) >sp_ag.0 1737268 120 - 2160267/0-120 AGGUCGGUGGUUCGAGUCCACUCGUGCCCAUCAAGAAUAUGGUCCGUUGGUCAAGGGGUUAAGACACCGCCUUUUCACGGCGGUAACACGGGUUCGAAUCCCGUACGGACUAUUCUUAAA .....(((((..((((....))))...)))))((((..(((((((((..(((..........)))((((((.......))))))...(((((.......))))))))))))))))))... ( -44.70) >consensus CGGUCGUAGGUUCGAGUCC_____UACCUGUGGAG_CCAUGGCCCCUUGGUCAAGCGGUUAAGACACCGCCCUUUCACGGCGGUAACACGGGUUCGAGUCCCGUAGGAGUCAUACAAACA ...((((((((..............)))))))).....(((((((((..(((..........)))((((((.......))))))...(((((.......))))))))))))))....... (-38.18 = -36.37 + -1.81) # Strand winner: reverse (0.99)

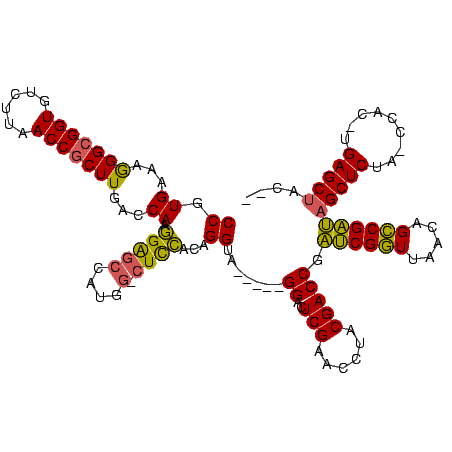
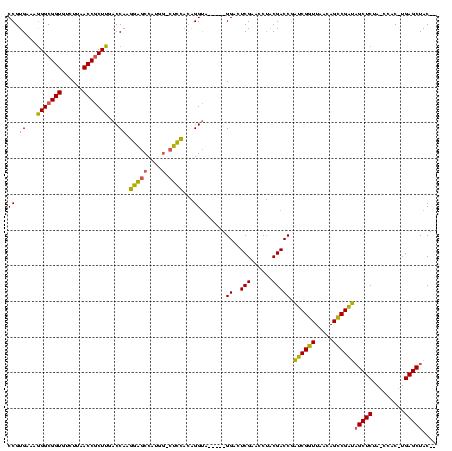
| Location | 1,940,108 – 1,940,218 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.20 |
| Mean single sequence MFE | -41.50 |
| Consensus MFE | -29.73 |
| Energy contribution | -28.60 |
| Covariance contribution | -1.13 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1940108 110 + 2809422/40-160 CCGUGAAAGGGCGGUGUCUUAACCGCUUGACCAAGGAGCCAUGG-CUCCACAGGUA-----GGACUCGAACCUACGACCGAUCGGUUAACAGCCGAUAGCUCUA-CCAC-UGAGCUAC-- ((......)).((((...(((((((.(((.....(((((....)-))))....(((-----((.......)))))...))).)))))))..)))).((((((..-....-.)))))).-- ( -41.60) >sy_sa.0 1512280 110 - 2516575/40-160 CCGUGAAAGGGCGGUGUCUUAACCGCUUGACCACGGGGCCAUGG-CUCCACAGGUA-----GGACUCGAACCUACGACCGAUCGGUUAACAGCCGAUAGCUCUA-CCAC-UGAGCUAC-- ..(((...(((((((......)))))))...)))(((((....)-))))....(((-----((.......))))).....((((((.....))))))(((((..-....-.)))))..-- ( -43.80) >sy_ha.0 1543851 110 - 2685015/40-160 CCGUGAAAGGGCGGUGUCUUAACCGCUUGACCAAGGAGCCAUGG-CUCCGCAGGUA-----GGAUUCGAACCUACGACCGAUCGGUUAACAGCCGAUAGCUCUA-CCAC-UGAGCUAC-- ((......)).((((...(((((((.(((.....(((((....)-))))....(((-----((.......)))))...))).)))))))..)))).((((((..-....-.)))))).-- ( -41.60) >sp_ag.0 1737268 120 - 2160267/40-160 CCGUGAAAAGGCGGUGUCUUAACCCCUUGACCAACGGACCAUAUUCUUGAUGGGCACGAGUGGACUCGAACCACCGACCUCACGCUUAUCAGGCGUGCGCUCUAACCACCUGAGCUACGC ((((...((((.(((......))))))).....)))).........(((((((((....((((..(((......)))..)))))))))))))(((((.((((.........))))))))) ( -39.00) >consensus CCGUGAAAGGGCGGUGUCUUAACCGCUUGACCAAGGAGCCAUGG_CUCCACAGGUA_____GGACUCGAACCUACGACCGAUCGGUUAACAGCCGAUAGCUCUA_CCAC_UGAGCUAC__ ((.((...(((((((......)))))))...)).(((((....).))))...)).......((..(((......))))).((((((.....))))))(((((.........))))).... (-29.73 = -28.60 + -1.13)

| Location | 1,940,108 – 1,940,201 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 67.56 |
| Mean single sequence MFE | -33.23 |
| Consensus MFE | -22.67 |
| Energy contribution | -20.55 |
| Covariance contribution | -2.12 |
| Combinations/Pair | 1.36 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.68 |
| SVM decision value | 5.24 |
| SVM RNA-class probability | 0.999980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1940108 93 + 2809422/80-200 AUGG-CUCCACAGGUA-----GGACUCGAACCUACGACCGAUCGGUUAACAGCCGAUAGCUCUA-CCAC-UGAGCUAC----------UGUGGAAUAAUAGAAAAU---------AAUGG ....-.((((((((((-----((.......))))).....((((((.....))))))(((((..-....-.))))).)----------))))))............---------..... ( -31.00) >sy_sa.0 1512280 89 - 2516575/80-200 AUGG-CUCCACAGGUA-----GGACUCGAACCUACGACCGAUCGGUUAACAGCCGAUAGCUCUA-CCAC-UGAGCUAC----------UGUGGAAUAAUUAAAA-------------UGG ....-.((((((((((-----((.......))))).....((((((.....))))))(((((..-....-.))))).)----------))))))..........-------------... ( -31.00) >sy_ha.0 1543851 88 - 2685015/80-200 AUGG-CUCCGCAGGUA-----GGAUUCGAACCUACGACCGAUCGGUUAACAGCCGAUAGCUCUA-CCAC-UGAGCUAC----------UGCGGAAUAAUAUGA--------------UGG ....-.((((((((((-----((.......))))).....((((((.....))))))(((((..-....-.))))).)----------)))))).........--------------... ( -32.50) >sp_ag.0 1737268 120 - 2160267/80-200 AUAUUCUUGAUGGGCACGAGUGGACUCGAACCACCGACCUCACGCUUAUCAGGCGUGCGCUCUAACCACCUGAGCUACGCGCCCAAGCUAUUGCUUGGUUUUUACUUUCUUAUAAAGUAA ......(((((((((....((((..(((......)))..)))))))))))))(((((.((((.........)))))))))..((((((....))))))...(((((((.....))))))) ( -38.40) >consensus AUGG_CUCCACAGGUA_____GGACUCGAACCUACGACCGAUCGGUUAACAGCCGAUAGCUCUA_CCAC_UGAGCUAC__________UGUGGAAUAAUAUAAA_____________UGG ......((((((.........((..(((......))))).((((((.....))))))(((((.........)))))............)))))).......................... (-22.67 = -20.55 + -2.12)

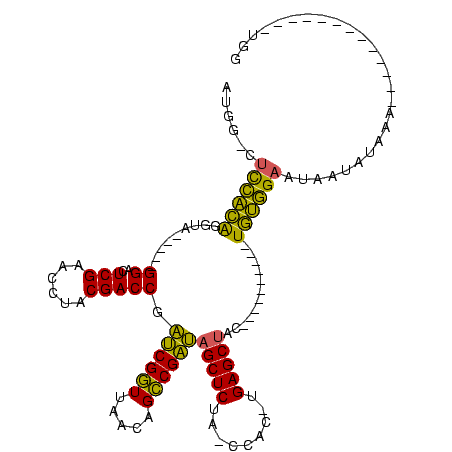
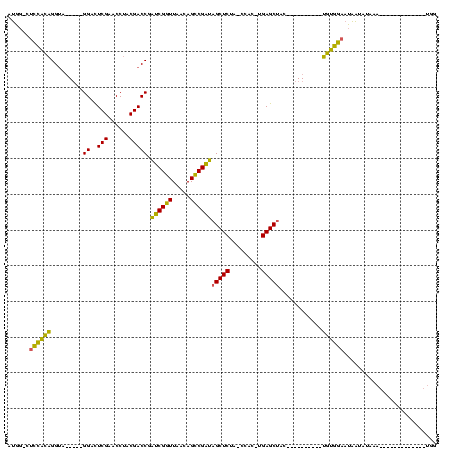
| Location | 1,940,108 – 1,940,201 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 67.56 |
| Mean single sequence MFE | -32.22 |
| Consensus MFE | -17.06 |
| Energy contribution | -15.75 |
| Covariance contribution | -1.31 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1940108 93 + 2809422/80-200 CCAUU---------AUUUUCUAUUAUUCCACA----------GUAGCUCA-GUGG-UAGAGCUAUCGGCUGUUAACCGAUCGGUCGUAGGUUCGAGUCC-----UACCUGUGGAG-CCAU .....---------...........(((((((----------(((((((.-....-..))))))..(((((.........)))))(((((.......))-----)))))))))))-.... ( -28.40) >sy_sa.0 1512280 89 - 2516575/80-200 CCA-------------UUUUAAUUAUUCCACA----------GUAGCUCA-GUGG-UAGAGCUAUCGGCUGUUAACCGAUCGGUCGUAGGUUCGAGUCC-----UACCUGUGGAG-CCAU ...-------------.........(((((((----------(((((((.-....-..))))))..(((((.........)))))(((((.......))-----)))))))))))-.... ( -28.40) >sy_ha.0 1543851 88 - 2685015/80-200 CCA--------------UCAUAUUAUUCCGCA----------GUAGCUCA-GUGG-UAGAGCUAUCGGCUGUUAACCGAUCGGUCGUAGGUUCGAAUCC-----UACCUGCGGAG-CCAU ...--------------........(((((((----------(((((((.-....-..))))))..(((((.........)))))(((((.......))-----)))))))))))-.... ( -29.90) >sp_ag.0 1737268 120 - 2160267/80-200 UUACUUUAUAAGAAAGUAAAAACCAAGCAAUAGCUUGGGCGCGUAGCUCAGGUGGUUAGAGCGCACGCCUGAUAAGCGUGAGGUCGGUGGUUCGAGUCCACUCGUGCCCAUCAAGAAUAU (((((((.....)))))))...((((((....)))))).(((((...(((((((((......)).)))))))...))))).....(((((..((((....))))...)))))........ ( -42.20) >consensus CCA_____________UUUAAAUUAUUCCACA__________GUAGCUCA_GUGG_UAGAGCUAUCGGCUGUUAACCGAUCGGUCGUAGGUUCGAGUCC_____UACCUGUGGAG_CCAU ..........................(((((..............((((.........)))).(((((.......))))).(((....((.......))......))).)))))...... (-17.06 = -15.75 + -1.31) # Strand winner: forward (0.96)

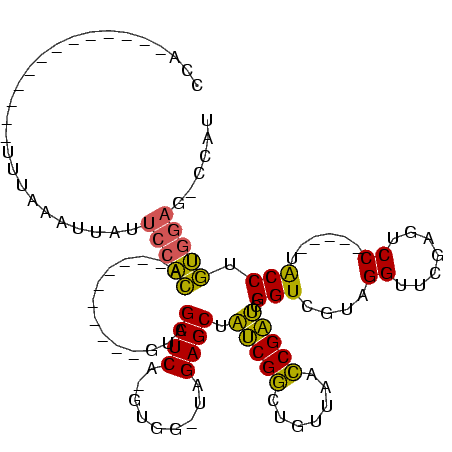
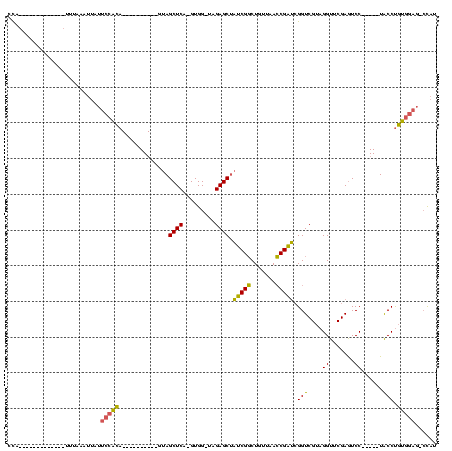
| Location | 1,940,108 – 1,940,207 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.97 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -25.26 |
| Energy contribution | -23.70 |
| Covariance contribution | -1.56 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.82 |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.995403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1940108 99 + 2809422/120-240 AUCGGUUAACAGCCGAUAGCUCUA-CCAC-UGAGCUAC----------UGUGGAAUAAUAGAAAAU---------AAUGGAGCGGGUGAUGGGAAUCGAACCCACAACAUCAGCUUGGAA .(((((.....)))))((((((..-....-.))))))(----------(((......)))).....---------..(.((((.((((.((((.......))))...)))).)))).).. ( -28.70) >sy_sa.0 1512280 95 - 2516575/120-240 AUCGGUUAACAGCCGAUAGCUCUA-CCAC-UGAGCUAC----------UGUGGAAUAAUUAAAA-------------UGGAGCGGGUGAUGGGAAUCGAACCCACAACAUCAGCUUGGAA .(((((.....)))))((((((..-....-.)))))).----------................-------------(.((((.((((.((((.......))))...)))).)))).).. ( -26.60) >sy_ha.0 1543851 94 - 2685015/120-240 AUCGGUUAACAGCCGAUAGCUCUA-CCAC-UGAGCUAC----------UGCGGAAUAAUAUGA--------------UGGAGCGGGUGAUGGGAAUCGAACCCACAACAUCAGCUUGGAA .(((((.....)))))((((((..-....-.)))))).----------............(((--------------((..(.(((((((....)))..)))).)..)))))........ ( -27.50) >sp_ag.0 1737268 120 - 2160267/120-240 CACGCUUAUCAGGCGUGCGCUCUAACCACCUGAGCUACGCGCCCAAGCUAUUGCUUGGUUUUUACUUUCUUAUAAAGUAAAGCGGGUGACGAGAAUCGAACUCGCGACAACAGCUUGGAA ((((((.....)))))).((((.........)))).......(((((((.((((......((((((((.....))))))))((((((..((.....)).))))))).))).))))))).. ( -40.40) >consensus AUCGGUUAACAGCCGAUAGCUCUA_CCAC_UGAGCUAC__________UGUGGAAUAAUAUAAA_____________UGGAGCGGGUGAUGGGAAUCGAACCCACAACAUCAGCUUGGAA ((((((.....))))))(((((.........)))))...........................................((((.((((.((((.......))))...)))).)))).... (-25.26 = -23.70 + -1.56)

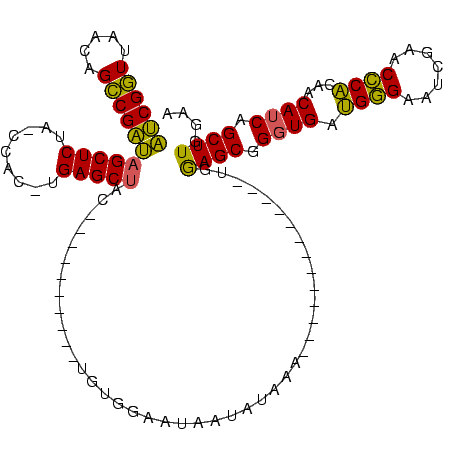
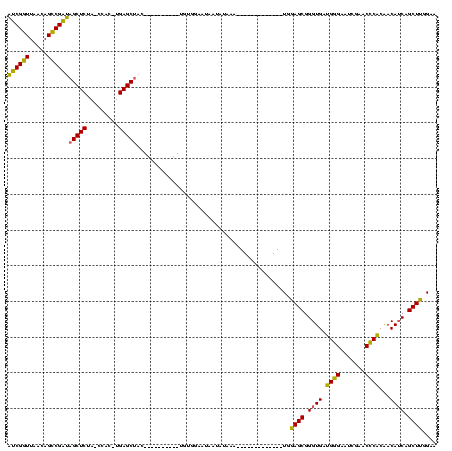
| Location | 1,940,108 – 1,940,207 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.97 |
| Mean single sequence MFE | -30.65 |
| Consensus MFE | -19.59 |
| Energy contribution | -19.65 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1940108 99 + 2809422/120-240 UUCCAAGCUGAUGUUGUGGGUUCGAUUCCCAUCACCCGCUCCAUU---------AUUUUCUAUUAUUCCACA----------GUAGCUCA-GUGG-UAGAGCUAUCGGCUGUUAACCGAU .....(((((((((.(((((.......))))).))..((((.(((---------(((..(((((.......)----------))))...)-))))-).)))).))))))).......... ( -25.80) >sy_sa.0 1512280 95 - 2516575/120-240 UUCCAAGCUGAUGUUGUGGGUUCGAUUCCCAUCACCCGCUCCA-------------UUUUAAUUAUUCCACA----------GUAGCUCA-GUGG-UAGAGCUAUCGGCUGUUAACCGAU .....((((((((..((((((..(((....)))))))))..))-------------)...............----------(((((((.-....-..)))))))))))).......... ( -24.80) >sy_ha.0 1543851 94 - 2685015/120-240 UUCCAAGCUGAUGUUGUGGGUUCGAUUCCCAUCACCCGCUCCA--------------UCAUAUUAUUCCGCA----------GUAGCUCA-GUGG-UAGAGCUAUCGGCUGUUAACCGAU .....((((((((..((((((..(((....)))))))))..))--------------)).............----------(((((((.-....-..))))))).)))).......... ( -26.80) >sp_ag.0 1737268 120 - 2160267/120-240 UUCCAAGCUGUUGUCGCGAGUUCGAUUCUCGUCACCCGCUUUACUUUAUAAGAAAGUAAAAACCAAGCAAUAGCUUGGGCGCGUAGCUCAGGUGGUUAGAGCGCACGCCUGAUAAGCGUG ..((((((((((((.(((.((.((.....))..)).)))((((((((.....))))))))......))))))))))))((((.....(((((((((......)).)))))))...)))). ( -45.20) >consensus UUCCAAGCUGAUGUUGUGGGUUCGAUUCCCAUCACCCGCUCCA_____________UUUAAAUUAUUCCACA__________GUAGCUCA_GUGG_UAGAGCUAUCGGCUGUUAACCGAU .....((((((((..((((((..(((....)))))))))..))........................................((((((.........)))))))))))).......... (-19.59 = -19.65 + 0.06) # Strand winner: forward (0.96)

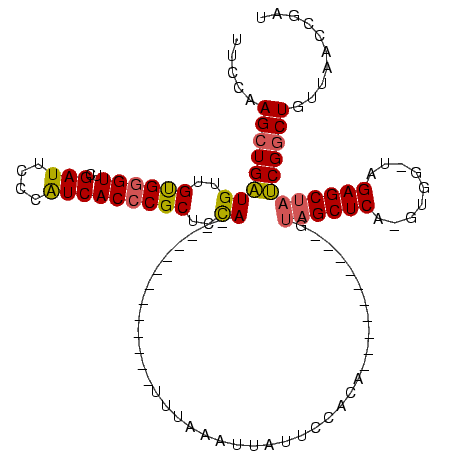
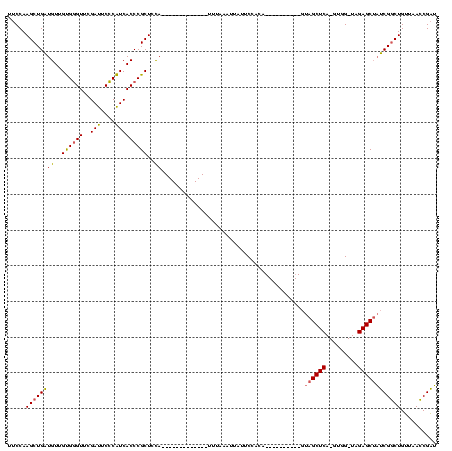
| Location | 1,940,108 – 1,940,211 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.36 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -28.89 |
| Energy contribution | -27.57 |
| Covariance contribution | -1.31 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.17 |
| SVM RNA-class probability | 0.999822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_au.0 1940108 103 + 2809422/160-280 --------UGUGGAAUAAUAGAAAAU---------AAUGGAGCGGGUGAUGGGAAUCGAACCCACAACAUCAGCUUGGAAGGCUGAGGUUUUGCCAUUAAACUACACCCGCUAAUGUAAA --------..................---------.....((((((((.((((.......)))).....((((((.....))))))((.....)).........))))))))........ ( -29.70) >sy_sa.0 1512280 97 - 2516575/160-280 --------UGUGGAAUAAUUAAAA-------------UGGAGCGGGUGAUGGGAAUCGAACCCACAACAUCAGCUUGGAAGGCUGAGGUUUUGCCAUUAAACUACACCCGC--ACUUUAA --------................-------------....(((((((.((((.......)))).....((((((.....))))))((.....)).........)))))))--....... ( -28.90) >sy_ha.0 1543851 97 - 2685015/160-280 --------UGCGGAAUAAUAUGA--------------UGGAGCGGGUGAUGGGAAUCGAACCCACAACAUCAGCUUGGAAGGCUGAGGUUUUGCCAUUAAACUACACCCGCGUA-GUAAA --------...............--------------....(((((((.((((.......)))).....((((((.....))))))((.....)).........)))))))...-..... ( -28.90) >sp_ag.0 1737268 111 - 2160267/160-280 GCCCAAGCUAUUGCUUGGUUUUUACUUUCUUAUAAAGUAAAGCGGGUGACGAGAAUCGAACUCGCGACAACAGCUUGGAAGGCUGUAGUUUUACCACUAAACUACACCCGC--------- ..((((((....))))))..((((((((.....))))))))(((((((.((((.......)))).(((.((((((.....)))))).)))..............)))))))--------- ( -39.30) >consensus ________UGUGGAAUAAUAUAAA_____________UGGAGCGGGUGAUGGGAAUCGAACCCACAACAUCAGCUUGGAAGGCUGAGGUUUUGCCAUUAAACUACACCCGC__A_GUAAA .........................................(((((((.((((.......)))).(((.((((((.....)))))).)))..............)))))))......... (-28.89 = -27.57 + -1.31)

| Location | 1,940,108 – 1,940,211 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.36 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -27.42 |
| Energy contribution | -26.55 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.86 |
| SVM decision value | 4.94 |
| SVM RNA-class probability | 0.999964 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
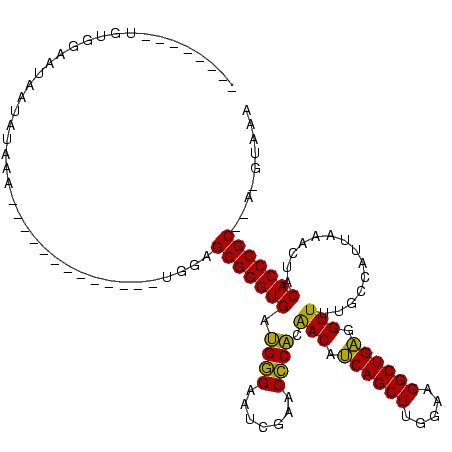
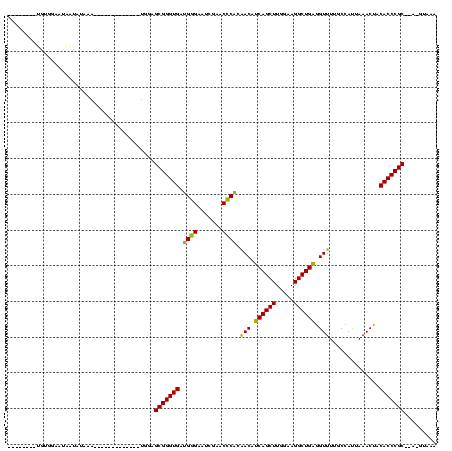
>sy_au.0 1940108 103 + 2809422/160-280 UUUACAUUAGCGGGUGUAGUUUAAUGGCAAAACCUCAGCCUUCCAAGCUGAUGUUGUGGGUUCGAUUCCCAUCACCCGCUCCAUU---------AUUUUCUAUUAUUCCACA-------- ........((((((((..............(((.(((((.......))))).)))(((((.......))))))))))))).....---------..................-------- ( -27.80) >sy_sa.0 1512280 97 - 2516575/160-280 UUAAAGU--GCGGGUGUAGUUUAAUGGCAAAACCUCAGCCUUCCAAGCUGAUGUUGUGGGUUCGAUUCCCAUCACCCGCUCCA-------------UUUUAAUUAUUCCACA-------- (((((((--(((((((..............(((.(((((.......))))).)))(((((.......))))))))))))...)-------------))))))..........-------- ( -28.30) >sy_ha.0 1543851 97 - 2685015/160-280 UUUAC-UACGCGGGUGUAGUUUAAUGGCAAAACCUCAGCCUUCCAAGCUGAUGUUGUGGGUUCGAUUCCCAUCACCCGCUCCA--------------UCAUAUUAUUCCGCA-------- .....-...(((((.(((((...((((...(((.(((((.......))))).)))((((((..(((....))))))))).)))--------------)...)))))))))).-------- ( -29.90) >sp_ag.0 1737268 111 - 2160267/160-280 ---------GCGGGUGUAGUUUAGUGGUAAAACUACAGCCUUCCAAGCUGUUGUCGCGAGUUCGAUUCUCGUCACCCGCUUUACUUUAUAAGAAAGUAAAAACCAAGCAAUAGCUUGGGC ---------((((.((((((((.......)))))))).))..((((((((((((.(((.((.((.....))..)).)))((((((((.....))))))))......)))))))))))))) ( -41.30) >consensus UUUAC_U__GCGGGUGUAGUUUAAUGGCAAAACCUCAGCCUUCCAAGCUGAUGUUGUGGGUUCGAUUCCCAUCACCCGCUCCA_____________UUUAAAUUAUUCCACA________ .........(((((((..............(((.(((((.......))))).)))(((((.......))))))))))))......................................... (-27.42 = -26.55 + -0.87) # Strand winner: forward (0.73)

| Location | 1,940,108 – 1,940,228 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.70 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -31.92 |
| Energy contribution | -30.80 |
| Covariance contribution | -1.12 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.858045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
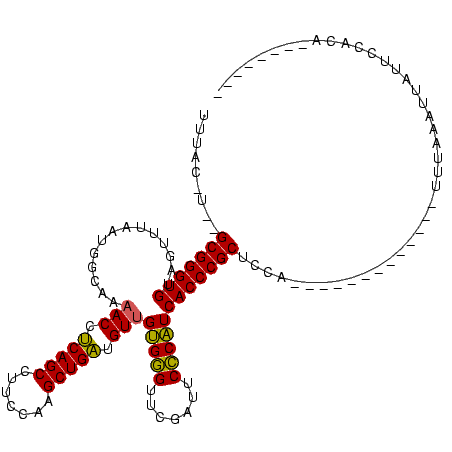
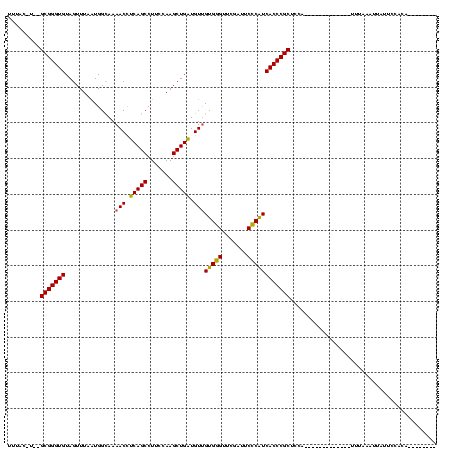
>sy_au.0 1940108 120 + 2809422/196-316 UCGAGGGUUCGAUCCCCUUCAGCCGCCCCAUAAUCGUUUACAUUAGCGGGUGUAGUUUAAUGGCAAAACCUCAGCCUUCCAAGCUGAUGUUGUGGGUUCGAUUCCCAUCACCCGCUCCAU ..(((((.((((.((((.((((((((((..((((.(....)))))..))))).........(((.........)))......)))))....).))).))))..))).))........... ( -33.70) >sy_sa.0 1512280 117 - 2516575/196-316 UCGUGGGUUCGAUUCCCAUCAUCCGCCCCAUAAUCGUUAAAGU--GCGGGUGUAGUUUAAUGGCAAAACCUCAGCCUUCCAAGCUGAUGUUGUGGGUUCGAUUCCCAUCACCCGCUCCA- ..((((((((((..(((((((((((((((((..........))--).))))).(((((...(((.........)))....)))))))))..))))).))))........))))))....- ( -36.30) >sy_ha.0 1543851 118 - 2685015/196-316 UCGAGGGUUCGAUUCCCUUCAACCGCCCCAUAAUCGUUUAC-UACGCGGGUGUAGUUUAAUGGCAAAACCUCAGCCUUCCAAGCUGAUGUUGUGGGUUCGAUUCCCAUCACCCGCUCCA- ..(((((........))))).....................-...(((((((..............(((.(((((.......))))).)))(((((.......))))))))))))....- ( -33.20) >sp_ag.0 1737268 107 - 2160267/196-316 UCAGCGGUUCGAU-CCCGUUAACUCCCAUAGAA------------GCGGGUGUAGUUUAGUGGUAAAACUACAGCCUUCCAAGCUGUUGUCGCGAGUUCGAUUCUCGUCACCCGCUUUAC ..(((((......-.)))))..........(((------------(((((((...............((.(((((.......))))).)).(((((.......))))))))))))))).. ( -32.80) >consensus UCGAGGGUUCGAUUCCCUUCAACCGCCCCAUAAUCGUUUAC_U__GCGGGUGUAGUUUAAUGGCAAAACCUCAGCCUUCCAAGCUGAUGUUGUGGGUUCGAUUCCCAUCACCCGCUCCA_ ..(((((........))))).........................(((((((..............(((.(((((.......))))).)))(((((.......))))))))))))..... (-31.92 = -30.80 + -1.12) # Strand winner: forward (0.92)

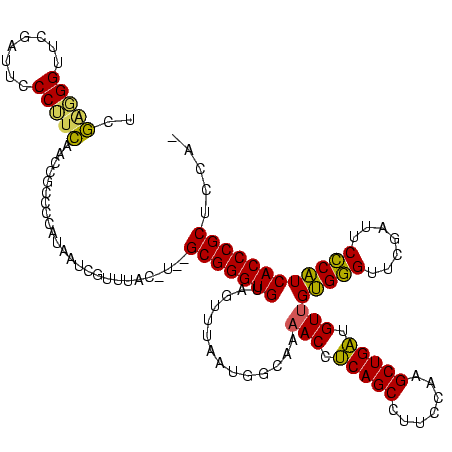
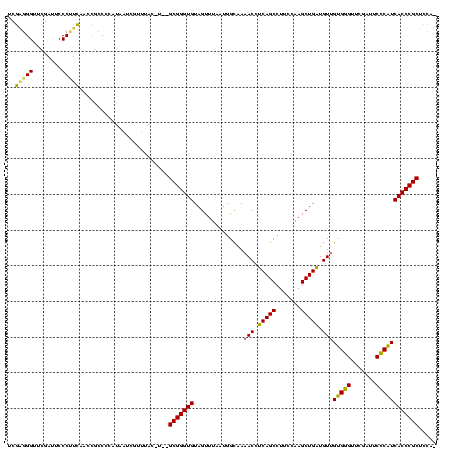
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:52:56 2006