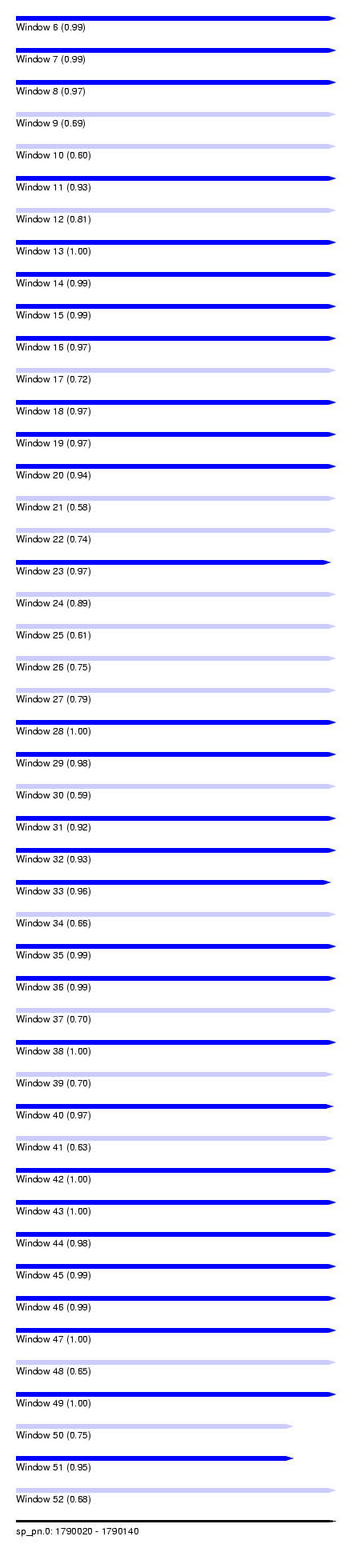
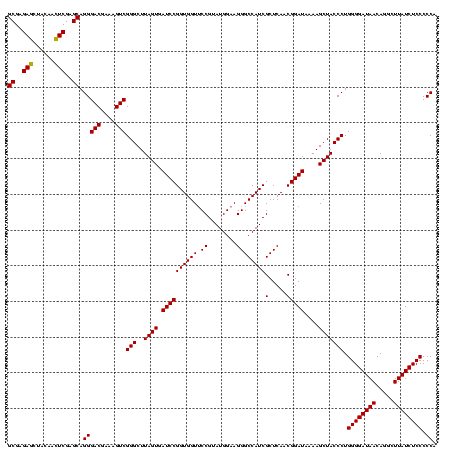
| Sequence ID | sp_pn.0 |
|---|---|
| Location | 1,790,020 – 1,790,140 |
| Length | 120 |
| Max. P | 0.998768 |

| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.06 |
| Mean single sequence MFE | -28.97 |
| Consensus MFE | -29.25 |
| Energy contribution | -27.70 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.15 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.992020 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/0-120 CAUGAUUAUAUAUCAGUAAGAGCCCUGAGAGAUGAUCAGGUAGAUAGGUUAGAAGUGGAAGUGUGGCGACACAUGUAGCGGACUAAUACUAAUAGCUCGAGGACUUAUCCAAAGUAACUG .............((((..(((((((((.......)))))....((((((((..((....(((((....)))))...))...))))).)))...))))..(((....)))......)))) ( -29.60) >sp_ag.0 2139123 120 - 2160267/0-120 CAUGAUUUUAUAUCAGUAAGAGCCCUGAGAGAUGAUCAGGUAGAUAGGUUAGGAGUGUAAUUGUGGUGACACAUUUAGCGGACUAAUACUAAUAGCUCGAGGACUUAUCCAAAAAGAGAU ..((((.....))))....(((((((((.......)))))....((((((((..((.(((.((((....)))).)))))...))))).)))...))))..(((....))).......... ( -29.50) >sp_mu.0 1684213 120 + 2030921/0-120 CAUAACGUUCAGUUAGUAAGAGCCCUGAGAGAAGAACAGGUAGAUAGGUUGGGAGUGGAAGCGUUGUGAGACGUGAAGCGGACCAAUACUAAUCGCUCGAGGACUUAUCCAAAAAUAAGU ..((((.....))))....((((((((.........))))..((((((((((..((....(((((....)))))...))...))))).)).)))))))....((((((......)))))) ( -27.80) >consensus CAUGAUUUUAUAUCAGUAAGAGCCCUGAGAGAUGAUCAGGUAGAUAGGUUAGGAGUGGAAGUGUGGUGACACAUGUAGCGGACUAAUACUAAUAGCUCGAGGACUUAUCCAAAAAAAAAU ..((((.....))))....(((((((((.......)))))....((((((((..((....(((((....)))))...))...))))).)))...))))..(((....))).......... (-29.25 = -27.70 + -1.55) # Strand winner: reverse (1.00)

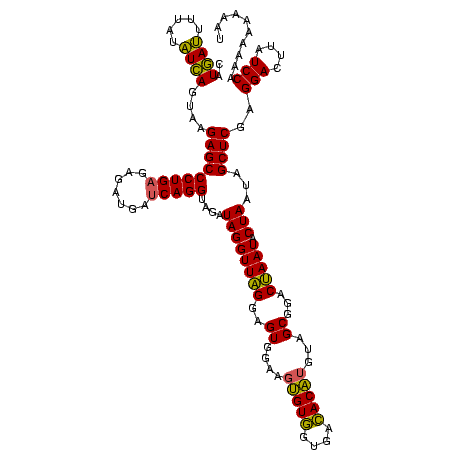

| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -60.70 |
| Consensus MFE | -60.70 |
| Energy contribution | -60.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.01 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/280-400 GCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGCGGGCGU (((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))..(((((.((((..(.(((((((....))).)))).)..))))..).)))))))).. ( -60.70) >sp_ag.0 2139123 120 - 2160267/280-400 GCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGCGGGCGU (((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))..(((((.((((..(.(((((((....))).)))).)..))))..).)))))))).. ( -60.70) >sp_mu.0 1684213 120 + 2030921/280-400 GCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGCGGGCGA (((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))..(((((.((((..(.(((((((....))).)))).)..))))..).)))))))).. ( -60.70) >consensus GCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGCGGGCGU (((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))..(((((.((((..(.(((((((....))).)))).)..))))..).)))))))).. (-60.70 = -60.70 + -0.00) # Strand winner: reverse (0.98)

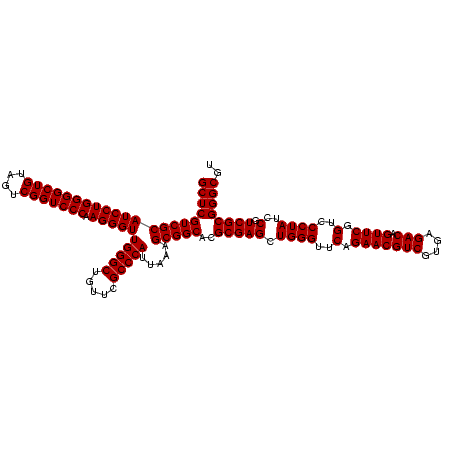
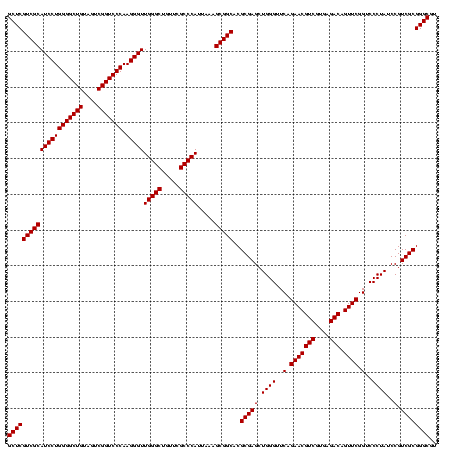
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -57.10 |
| Consensus MFE | -57.10 |
| Energy contribution | -57.10 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.55 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/320-440 AGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGGUUCAG .(((..((...(((((..(((((.....)))))..)))))(((((((((((((((((((((....))))))).)))))(((((.....))))).....)))....))))))))..))).. ( -57.10) >sp_ag.0 2139123 120 - 2160267/320-440 AGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGGUUCAG .(((..((...(((((..(((((.....)))))..)))))(((((((((((((((((((((....))))))).)))))(((((.....))))).....)))....))))))))..))).. ( -57.10) >sp_mu.0 1684213 120 + 2030921/320-440 AGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGGUUCAG .(((..((...(((((..(((((.....)))))..)))))(((((((((((((((((((((....))))))).)))))(((((.....))))).....)))....))))))))..))).. ( -57.10) >consensus AGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGGUUCAG .(((..((...(((((..(((((.....)))))..)))))(((((((((((((((((((((....))))))).)))))(((((.....))))).....)))....))))))))..))).. (-57.10 = -57.10 + 0.00) # Strand winner: reverse (1.00)

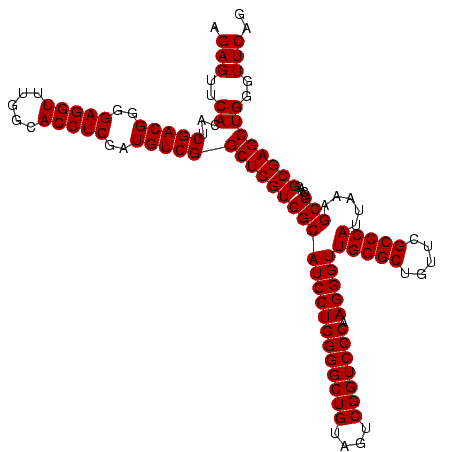
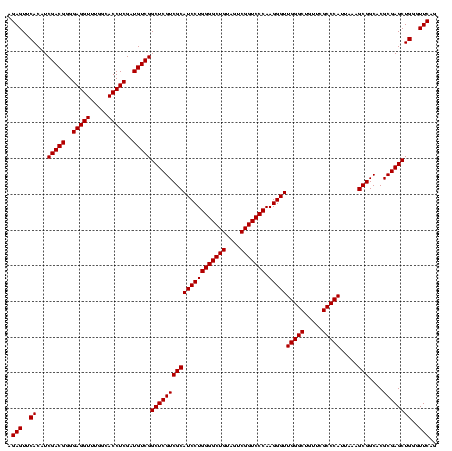
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -47.50 |
| Consensus MFE | -47.50 |
| Energy contribution | -47.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/360-480 GAUAAAAGCUACCCUGGGGAUAACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUG ..........(((((.(((((.(((((((.((((((((...((.......))...))))))))..))).((((((((.(((....)))))))..)))))))).....))))).))))).. ( -47.50) >sp_ag.0 2139123 120 - 2160267/360-480 GAUAAAAGCUACCCUGGGGAUAACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUG ..........(((((.(((((.(((((((.((((((((...((.......))...))))))))..))).((((((((.(((....)))))))..)))))))).....))))).))))).. ( -47.50) >sp_mu.0 1684213 120 + 2030921/360-480 GAUAAAAGCUACCCUGGGGAUAACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUG ..........(((((.(((((.(((((((.((((((((...((.......))...))))))))..))).((((((((.(((....)))))))..)))))))).....))))).))))).. ( -47.50) >consensus GAUAAAAGCUACCCUGGGGAUAACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUG ..........(((((.(((((.(((((((.((((((((...((.......))...))))))))..))).((((((((.(((....)))))))..)))))))).....))))).))))).. (-47.50 = -47.50 + 0.00) # Strand winner: reverse (0.98)

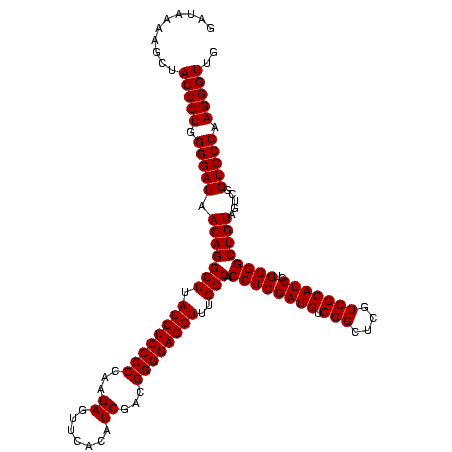
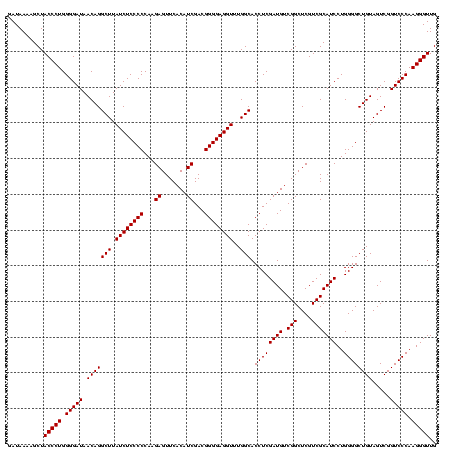
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -42.90 |
| Consensus MFE | -42.61 |
| Energy contribution | -42.17 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601466 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/440-560 GCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGCUACCCUGGGGAUAACAGGCUUAUCUCCCCCA ((..(((......)))..)).(((((....)))(((..((((.(((((....((((....))))((((....))))..)))))....))))))).((((((((.....)))))))).)). ( -43.10) >sp_ag.0 2139123 120 - 2160267/440-560 GCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGCUACCCUGGGGAUAACAGGCUUAUCUCCCCCA ((..(((......)))..)).(((((....)))(((..((((.(((((....((((....))))((((....))))..)))))....))))))).((((((((.....)))))))).)). ( -43.10) >sp_mu.0 1684213 120 + 2030921/440-560 GCGAGACAGACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUACCGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGCUACCCUGGGGAUAACAGGCUUAUCUCCCCCA ((..(((......)))..)).(((((....)))(((..((((.((((((((((.((........))))))))(.....)))))....))))))).((((((((.....)))))))).)). ( -42.50) >consensus GCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGUAUGGAAGGGCCAUCGCUCAACGGAUAAAAGCUACCCUGGGGAUAACAGGCUUAUCUCCCCCA ((..(((......)))..)).(((((....)))(((..((((.((((((((((.((........))))))))(.....)))))....))))))).((((((((.....)))))))).)). (-42.61 = -42.17 + -0.44) # Strand winner: reverse (0.96)


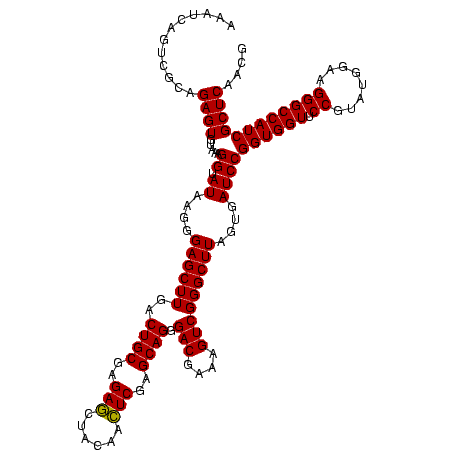
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -40.83 |
| Consensus MFE | -40.71 |
| Energy contribution | -40.27 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.00 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/480-600 AAAUCAUUCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGUAUGGAAGGGCCAUCGCUCAACG ............((((.....((.((....((((((..((((..(((......)))..)))).(((....)))))))))....))))((((((.((........)))))))))))).... ( -40.10) >sp_ag.0 2139123 120 - 2160267/480-600 AAAUCAGUCGUAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGUAUGGAAGGGCCAUCGCUCAACG ........(((.((((.....((.((....((((((..((((..(((......)))..)))).(((....)))))))))....))))((((((.((........)))))))))))).))) ( -41.80) >sp_mu.0 1684213 120 + 2030921/480-600 AAAUCAGUCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGACAGACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUACCGUAUGGAAGGGCCAUCGCUCAACG ............((((.....((.((....((((((..((((..(((......)))..)))).(((....)))))))))....))))((((((.((........)))))))))))).... ( -40.60) >consensus AAAUCAGUCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGUAUGGAAGGGCCAUCGCUCAACG ............((((.....((.((....((((((..((((..(((......)))..)))).(((....)))))))))....))))((((((.((........)))))))))))).... (-40.71 = -40.27 + -0.44) # Strand winner: reverse (1.00)

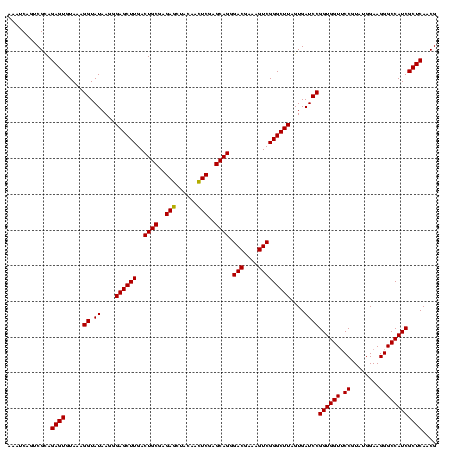
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -30.60 |
| Consensus MFE | -30.48 |
| Energy contribution | -29.59 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.65 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/520-640 UAAGCCCGACUUUCGUCCCUGCUCGAGUUGUAGCUCUCGCAGUCAAGCUCCCUUAUACCUUUACACUCUGCGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCU ...((((((.........((((..((((....))))..))))......(((((((........(.....).(((((.........))))))))))))....))))))............. ( -28.90) >sp_ag.0 2139123 120 - 2160267/520-640 UAAGCCCGACUUUCGUCCCUGCUCGAGUUGUAGCUCUCGCAGUCAAGCUCCCUUAUACCUUUACACUCUACGACUGAUUUCCAACCAGUCUGAGGGAACCUUUGGGCGCCUCCGUUACCU ...((((((.........((((..((((....))))..))))......(((((((................(((((.........))))))))))))....))))))............. ( -31.09) >sp_mu.0 1684213 120 + 2030921/520-640 UAAGCCCGACUUUCGUCCCUGCUCGACUUGUCUGUCUCGCAGUCAAGCUCCCUUAUACCUUUACACUCUGCGACUGAUUUCCAACCAGUCUGAGGGAACCUUUGGGCGCCUCCGUUACAC ...((((((......(((((....((((((.(((.....))).)))).)).....................(((((.........)))))..)))))....))))))............. ( -31.80) >consensus UAAGCCCGACUUUCGUCCCUGCUCGAGUUGUAGCUCUCGCAGUCAAGCUCCCUUAUACCUUUACACUCUGCGACUGAUUUCCAACCAGUCUGAGGGAACCUUUGGGCGCCUCCGUUACCU ...((((((.........((((..(((......)))..))))......(((((((................(((((.........))))))))))))....))))))............. (-30.48 = -29.59 + -0.89)

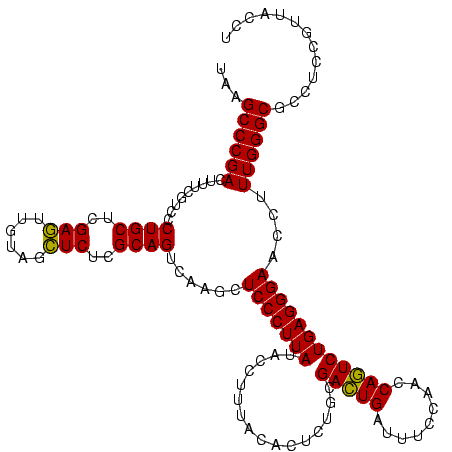

| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -44.20 |
| Consensus MFE | -44.89 |
| Energy contribution | -44.00 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.34 |
| Structure conservation index | 1.02 |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.997071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/520-640 AGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGGGCUUA .............((((..(((((((((..(((((((.....)))))))((.....)).........)))))))))..((((..(((......)))..)))).(((....)))))))... ( -42.60) >sp_ag.0 2139123 120 - 2160267/520-640 AGGUAACGGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGGAAAUCAGUCGUAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGGGCUUA .............((((..(((((((((..(((((((.....)))))))......((((....)))))))))))))..((((..(((......)))..)))).(((....)))))))... ( -44.60) >sp_mu.0 1684213 120 + 2030921/520-640 GUGUAACGGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGGAAAUCAGUCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGACAGACAAGUCGAGCAGGGACGAAAGUCGGGCUUA .............((((..(((((((((..(((((((.....)))))))((.....)).........)))))))))..((((..(((......)))..)))).(((....)))))))... ( -45.40) >consensus AGGUAACGGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGGAAAUCAGUCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACUGCGAGAGCUACAACUCGAGCAGGGACGAAAGUCGGGCUUA .............((((..(((((((((..(((((((.....)))))))......((((....)))))))))))))..((((..(((......)))..)))).(((....)))))))... (-44.89 = -44.00 + -0.89) # Strand winner: reverse (1.00)

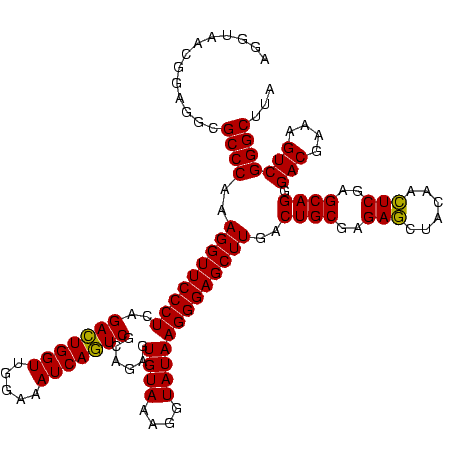
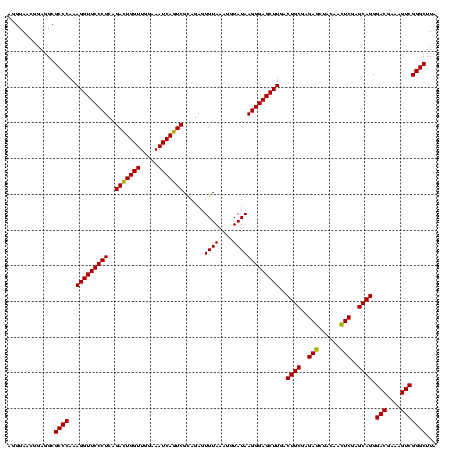
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -38.00 |
| Consensus MFE | -38.37 |
| Energy contribution | -37.92 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.01 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/560-680 AGUCAAGCUCCCUUAUACCUUUACACUCUGCGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACA .(((..(((((((((........(.....).(((((.........))))))))))))......((((((((((...........))))))(((.......))).....))))))..))). ( -36.50) >sp_ag.0 2139123 120 - 2160267/560-680 AGUCAAGCUCCCUUAUACCUUUACACUCUACGACUGAUUUCCAACCAGUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACA .(((..(((((((((................(((((.........))))))))))))......((((((((((...........))))))(((.......))).....))))))..))). ( -38.69) >sp_mu.0 1684213 120 + 2030921/560-680 AGUCAAGCUCCCUUAUACCUUUACACUCUGCGACUGAUUUCCAACCAGUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACA .(((..(((((((((........(.....).(((((.........))))))))))))......((((((((((...........))))))(((.......))).....))))))..))). ( -38.80) >consensus AGUCAAGCUCCCUUAUACCUUUACACUCUGCGACUGAUUUCCAACCAGUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACA .(((..(((((((((................(((((.........))))))))))))......((((((((((...........))))))(((.......))).....))))))..))). (-38.37 = -37.92 + -0.44)

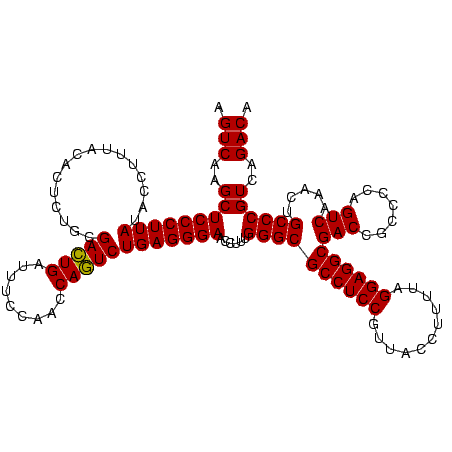
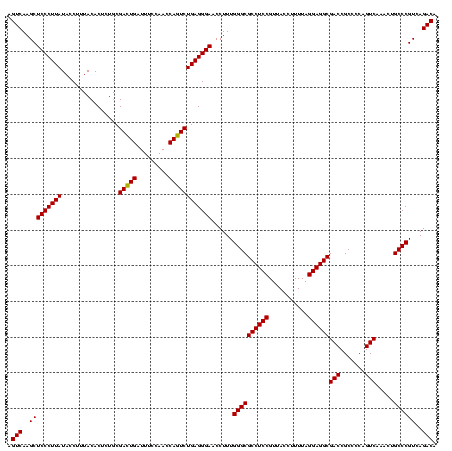
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -46.53 |
| Consensus MFE | -46.78 |
| Energy contribution | -46.33 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.68 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/560-680 UGUCUGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACU .(((....((((.....(((((...)))))((((((...........))))))))))..(((((((((..(((((((.....)))))))((.....)).........)))))))))))). ( -45.10) >sp_ag.0 2139123 120 - 2160267/560-680 UGUCUGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGGAAAUCAGUCGUAGAGUGUAAAGGUAUAAGGGAGCUUGACU .(((....((((.....(((((...)))))((((((...........))))))))))..(((((((((..(((((((.....)))))))......((((....)))))))))))))))). ( -47.10) >sp_mu.0 1684213 120 + 2030921/560-680 UGUCUGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGGAAAUCAGUCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACU .(((....((((.....(((((...)))))((((((...........))))))))))..(((((((((..(((((((.....)))))))((.....)).........)))))))))))). ( -47.40) >consensus UGUCUGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGGAAAUCAGUCGCAGAGUGUAAAGGUAUAAGGGAGCUUGACU .(((....((((.....(((((...)))))((((((...........))))))))))..(((((((((..(((((((.....)))))))......((((....)))))))))))))))). (-46.78 = -46.33 + -0.44) # Strand winner: reverse (1.00)

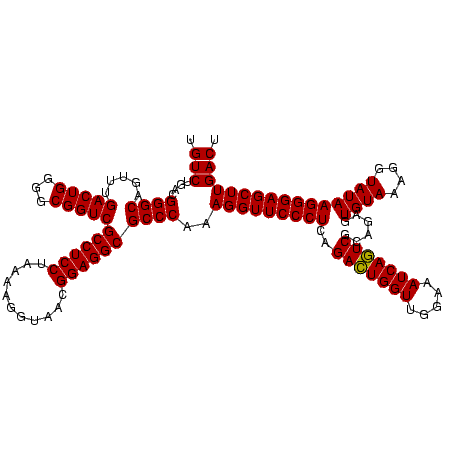
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -44.27 |
| Consensus MFE | -44.37 |
| Energy contribution | -43.27 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.94 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.967672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/600-720 CCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCGAUAGGGAUCACCUAUCUGGGUUAGAGUGGCC ....((((((((((((.......((((((((((...........))))))....))))(((.....)))))).)))))..(((.(((.((((((.....)))))).))).))).)))).. ( -46.10) >sp_ag.0 2139123 120 - 2160267/600-720 CCAACCAGUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCGAUAGGGAUUGCCUAUCUGGGUUAGAGUAGCC .......(((((((((.......((((((((((...........))))))....))))(((.....)))))).)))))).(((.(((.((((((.....)))))).))).)))....... ( -46.00) >sp_mu.0 1684213 120 + 2030921/600-720 CCAACCAGUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCGUAGAUGAUCAACCUACCGGGUUAGAGUAGCC .......(((((((((.......((((((((((...........))))))....))))(((.....)))))).)))))).(((.(((.((((.........)))).))).)))....... ( -40.70) >consensus CCAACCAGUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCGAUAGGGAUCACCUAUCUGGGUUAGAGUAGCC .......(((((((((.......((((((((((...........))))))....))))(((.....)))))).)))))).(((.(((.((((((.....)))))).))).)))....... (-44.37 = -43.27 + -1.11)

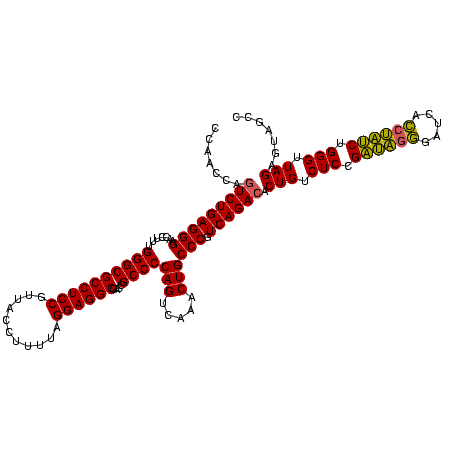
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -45.30 |
| Consensus MFE | -46.63 |
| Energy contribution | -44.97 |
| Covariance contribution | -1.66 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.07 |
| Structure conservation index | 1.03 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/600-720 GGCCACUCUAACCCAGAUAGGUGAUCCCUAUCGGAGACAGUGUCUGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGG .......((((((..((((((.....))))))(....)....(((((.((((((.....)))((((....((((((...........)))))))))).......))))))))..)))))) ( -44.40) >sp_ag.0 2139123 120 - 2160267/600-720 GGCUACUCUAACCCAGAUAGGCAAUCCCUAUCGGAGACAGUGUCUGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGG .......((((((..((((((.....))))))(....)...((((((.((((((.....)))((((....((((((...........)))))))))).......))))))))).)))))) ( -47.00) >sp_mu.0 1684213 120 + 2030921/600-720 GGCUACUCUAACCCGGUAGGUUGAUCAUCUACGGAGACAGUGUCUGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGG .......((((((..((((((.....))))))(....)...((((((.((((((.....)))((((....((((((...........)))))))))).......))))))))).)))))) ( -44.50) >consensus GGCUACUCUAACCCAGAUAGGUGAUCCCUAUCGGAGACAGUGUCUGACGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGACUGGUUGG .......((((((..((((((.....))))))(....)...((((((.((((((.....)))((((....((((((...........)))))))))).......))))))))).)))))) (-46.63 = -44.97 + -1.66) # Strand winner: reverse (0.96)

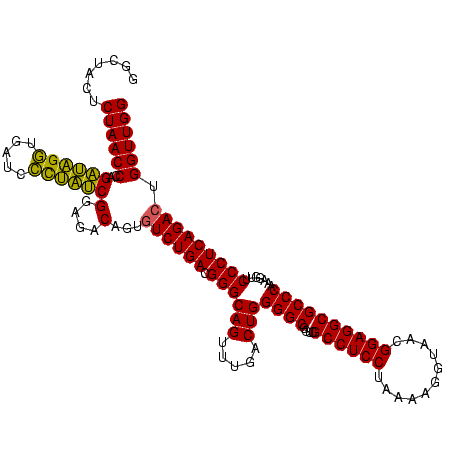
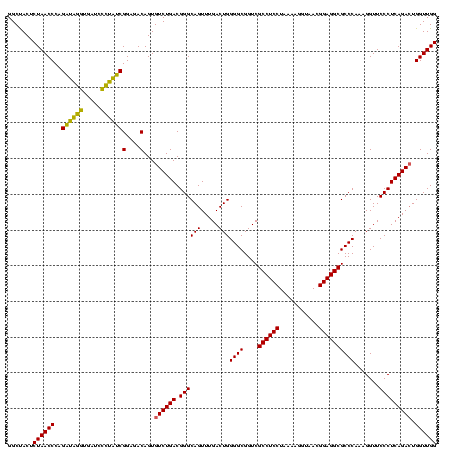
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.33 |
| Mean single sequence MFE | -37.87 |
| Consensus MFE | -36.59 |
| Energy contribution | -35.27 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/640-760 UUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCGAUAGGGAUCACCUAUCUGGGUUAGAGUGGCCAUAACACAAGGGUAGUAUCCCAACAGCGUCUCCUUCGAAA ...((((((((((.......)))..(((((((........(((.(((.((((((.....)))))).))).))).(((.......)))..)))))))...........)))))))...... ( -41.50) >sp_ag.0 2139123 120 - 2160267/640-760 UUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCGAUAGGGAUUGCCUAUCUGGGUUAGAGUAGCCAUAACACAAGGGUAGUAUCCCAACAACGCCUCAAACGAAA .....((((((((.......)))..(((((((........(((.(((.((((((.....)))))).))).))).((.......))....)))))))...........)))))........ ( -35.70) >sp_mu.0 1684213 120 + 2030921/640-760 UUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCGUAGAUGAUCAACCUACCGGGUUAGAGUAGCCAUCACACAAGGGUAGUAUCCCAACAACGCCUCCAAGUAAA ....(((((((((.......)))..(((((((...((((...))))..((.((((.((((((.....)))).)).....))))))....)))))))...........))))))....... ( -36.40) >consensus UUUAGGAGGCGACCGCCCCAGUCAAACUGCCCGUCAGACACUGUCUCCGAUAGGGAUCACCUAUCUGGGUUAGAGUAGCCAUAACACAAGGGUAGUAUCCCAACAACGCCUCCAACGAAA ....(((((((((.......)))..(((((((........(((.(((.((((((.....)))))).))).))).((.......))....)))))))...........))))))....... (-36.59 = -35.27 + -1.33)

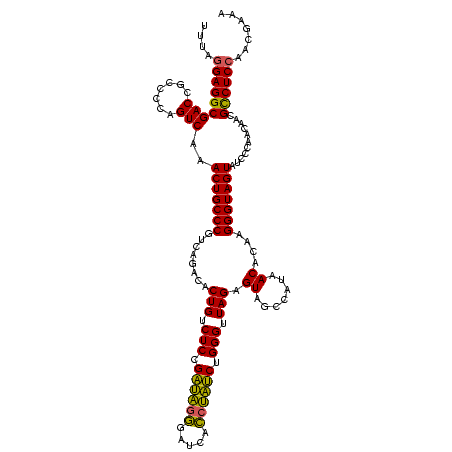
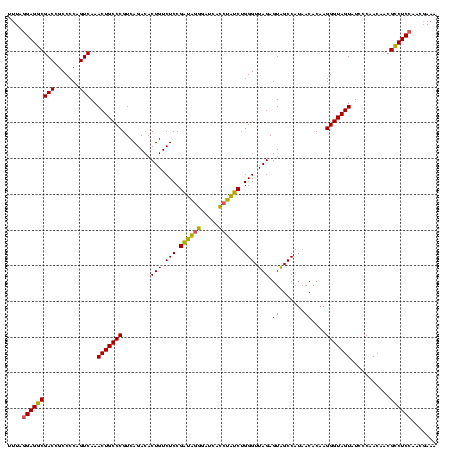
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -28.78 |
| Consensus MFE | -28.42 |
| Energy contribution | -25.10 |
| Covariance contribution | -3.32 |
| Combinations/Pair | 1.34 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/680-800 CUGUCUCCGAUAGGGAUCACCUAUCUGGGUUAGAGUGGCCAUAACACAAGGGUAGUAUCCCAACAGCGUCUCCUUCGAAACUGGCGUCCCGAUCUCUUAGACUCCUACCUAUCCUGUACA (((.(((.((((((.....)))))).))).))).(((.......))).(((((((........(((..((......))..)))..(((...........)))......)))))))..... ( -28.80) >sp_ag.0 2139123 120 - 2160267/680-800 CUGUCUCCGAUAGGGAUUGCCUAUCUGGGUUAGAGUAGCCAUAACACAAGGGUAGUAUCCCAACAACGCCUCAAACGAAACUGGCGUCCCGUUAUCAUAGGCUCCUACCUAUCCUGUACA ........((((((....((((((...(((((...)))))(((((....(((......)))....(((((............)))))...))))).)))))).....))))))....... ( -33.80) >sp_mu.0 1684213 120 + 2030921/680-800 CUGUCUCCGUAGAUGAUCAACCUACCGGGUUAGAGUAGCCAUCACACAAGGGUAGUAUCCCAACAACGCCUCCAAGUAAACUAGCGUUCACUCUUCCUUGGCUCCUACCUAUCCUGUACA ........((((.........))))((((.(((((.(((((........(((......)))...(((((..............)))))..........))))).))..))).)))).... ( -23.74) >consensus CUGUCUCCGAUAGGGAUCACCUAUCUGGGUUAGAGUAGCCAUAACACAAGGGUAGUAUCCCAACAACGCCUCCAACGAAACUGGCGUCCCGUUAUCAUAGGCUCCUACCUAUCCUGUACA ........((((((.....))))))((((.(((.((((...........(((......)))......((((...(((..((....))..)))......))))..))))))).)))).... (-28.42 = -25.10 + -3.32)

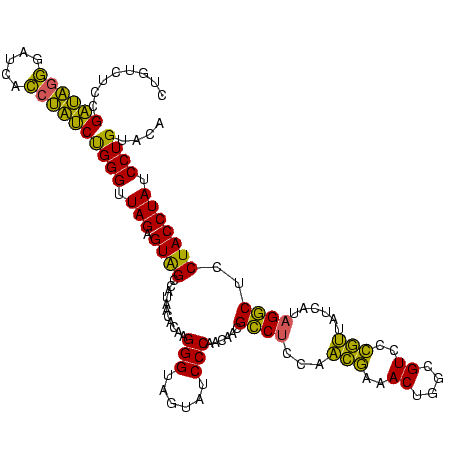
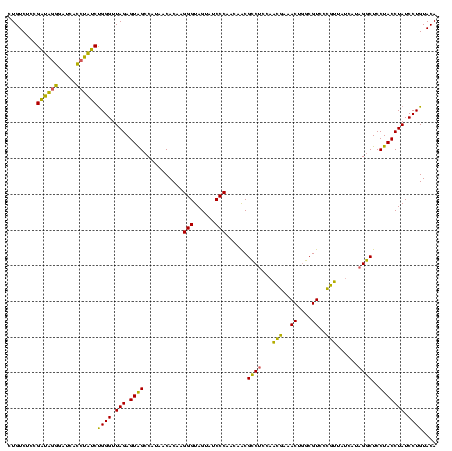
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -35.23 |
| Consensus MFE | -35.10 |
| Energy contribution | -32.00 |
| Covariance contribution | -3.10 |
| Combinations/Pair | 1.35 |
| Mean z-score | -0.27 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/680-800 UGUACAGGAUAGGUAGGAGUCUAAGAGAUCGGGACGCCAGUUUCGAAGGAGACGCUGUUGGGAUACUACCCUUGUGUUAUGGCCACUCUAACCCAGAUAGGUGAUCCCUAUCGGAGACAG ..((((((...(((((...(((((.((.(((..((....))..))).(....).)).)))))...)))))))))))...(((..........)))((((((.....))))))(....).. ( -37.10) >sp_ag.0 2139123 120 - 2160267/680-800 UGUACAGGAUAGGUAGGAGCCUAUGAUAACGGGACGCCAGUUUCGUUUGAGGCGUUGUUGGGAUACUACCCUUGUGUUAUGGCUACUCUAACCCAGAUAGGCAAUCCCUAUCGGAGACAG ......((.(((((((..((((.....((((..((....))..))))..))))......(((......)))...........)))).))).))..((((((.....))))))(....).. ( -36.30) >sp_mu.0 1684213 120 + 2030921/680-800 UGUACAGGAUAGGUAGGAGCCAAGGAAGAGUGAACGCUAGUUUACUUGGAGGCGUUGUUGGGAUACUACCCUUGUGUGAUGGCUACUCUAACCCGGUAGGUUGAUCAUCUACGGAGACAG ....(.((.(((((((..(((......((((((((....))))))))...)))((..(.(((......)))....)..))..)))).))).)).)((((((.....))))))(....).. ( -32.30) >consensus UGUACAGGAUAGGUAGGAGCCUAAGAAAACGGGACGCCAGUUUCGUUGGAGGCGUUGUUGGGAUACUACCCUUGUGUUAUGGCUACUCUAACCCAGAUAGGUGAUCCCUAUCGGAGACAG ......((.(((((((..((((.....((((((((....))))))))..))))......(((......)))...........)))).))).))..((((((.....))))))(....).. (-35.10 = -32.00 + -3.10) # Strand winner: reverse (1.00)

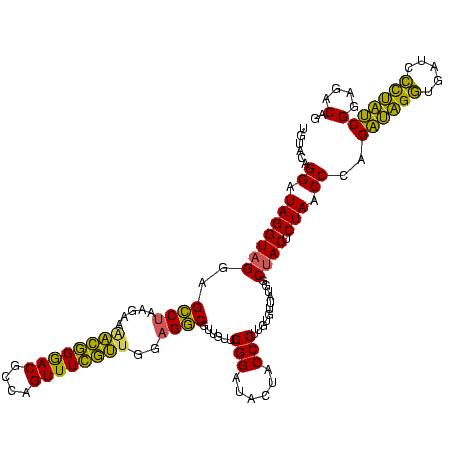
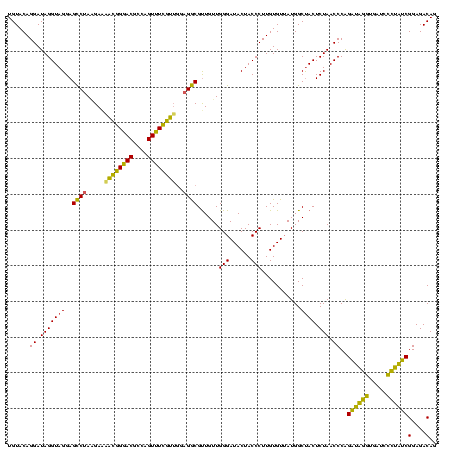
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -37.20 |
| Consensus MFE | -37.20 |
| Energy contribution | -37.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.38 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.577300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/840-960 AUGGGGUCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAAAAUUUCACCGAGUCUCUCGUUGAGACAGUGCCCAAAUCAUUACGCCUUUCGUGCGGGUCGGAACUUACC ....(((..(((((.((((.(((((((((((((........)))))................(((((.....)))))..)))))..........(.....)))))))).)))))...))) ( -37.20) >sp_ag.0 2139123 120 - 2160267/840-960 AUGGGGUCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAAAAUUUCACCGAGUCUCUCGUUGAGACAGUGCCCAAAUCAUUACGCCUUUCGUGCGGGUCGGAACUUACC ....(((..(((((.((((.(((((((((((((........)))))................(((((.....)))))..)))))..........(.....)))))))).)))))...))) ( -37.20) >sp_mu.0 1684213 120 + 2030921/840-960 AUGGGGUCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAAAAUUUCACCGAGUCUCUCGUUGAGACAGUGCCCAAAUCAUUACGCCUUUCGUGCGGGUCGGAACUUACC ....(((..(((((.((((.(((((((((((((........)))))................(((((.....)))))..)))))..........(.....)))))))).)))))...))) ( -37.20) >consensus AUGGGGUCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAAAAUUUCACCGAGUCUCUCGUUGAGACAGUGCCCAAAUCAUUACGCCUUUCGUGCGGGUCGGAACUUACC ....(((..(((((.((((.(((((((((((((........)))))................(((((.....)))))..)))))..........(.....)))))))).)))))...))) (-37.20 = -37.20 + -0.00)

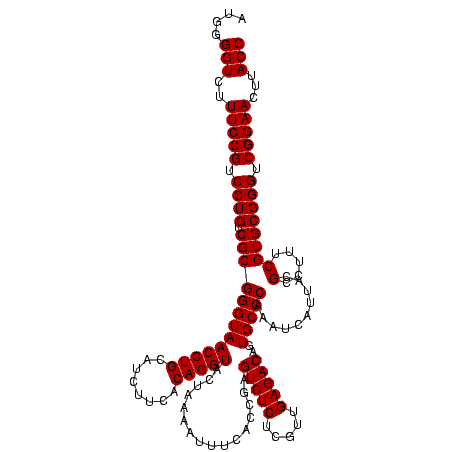
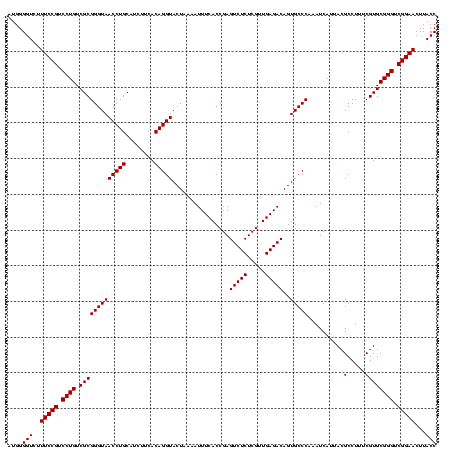
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -39.80 |
| Consensus MFE | -39.80 |
| Energy contribution | -39.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.61 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736278 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/840-960 GGUAAGUUCCGACCCGCACGAAAGGCGUAAUGAUUUGGGCACUGUCUCAACGAGAGACUCGGUGAAAUUUUAGUACCUGUGAAGAUGCAGGUUACCCGCGACAGGACGGAAAGACCCCAU (((...(((((.(((((.(....)))).........((((((((((((.....)))))..))))..........((((((......))))))..)))......)).)))))..))).... ( -39.80) >sp_ag.0 2139123 120 - 2160267/840-960 GGUAAGUUCCGACCCGCACGAAAGGCGUAAUGAUUUGGGCACUGUCUCAACGAGAGACUCGGUGAAAUUUUAGUACCUGUGAAGAUGCAGGUUACCCGCGACAGGACGGAAAGACCCCAU (((...(((((.(((((.(....)))).........((((((((((((.....)))))..))))..........((((((......))))))..)))......)).)))))..))).... ( -39.80) >sp_mu.0 1684213 120 + 2030921/840-960 GGUAAGUUCCGACCCGCACGAAAGGCGUAAUGAUUUGGGCACUGUCUCAACGAGAGACUCGGUGAAAUUUUAGUACCUGUGAAGAUGCAGGUUACCCGCGACAGGACGGAAAGACCCCAU (((...(((((.(((((.(....)))).........((((((((((((.....)))))..))))..........((((((......))))))..)))......)).)))))..))).... ( -39.80) >consensus GGUAAGUUCCGACCCGCACGAAAGGCGUAAUGAUUUGGGCACUGUCUCAACGAGAGACUCGGUGAAAUUUUAGUACCUGUGAAGAUGCAGGUUACCCGCGACAGGACGGAAAGACCCCAU (((...(((((.(((((.(....)))).........((((((((((((.....)))))..))))..........((((((......))))))..)))......)).)))))..))).... (-39.80 = -39.80 + 0.00) # Strand winner: reverse (0.90)

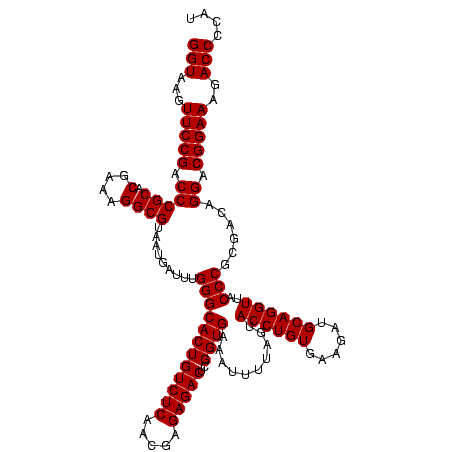
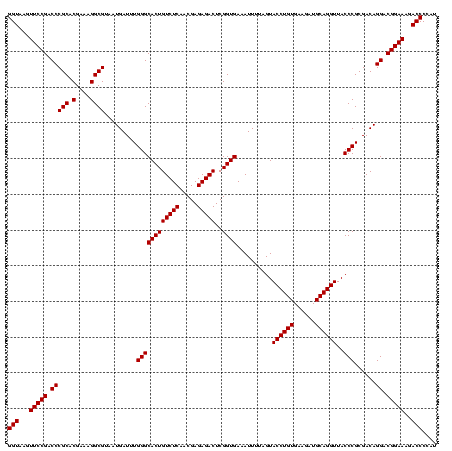
| Location | 1,790,020 – 1,790,138 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.53 |
| Mean single sequence MFE | -43.70 |
| Consensus MFE | -41.71 |
| Energy contribution | -41.43 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 118 + 2038615/1000-1120 AGCUCUCUGCUAAAUCGUAAGAUGAUGUAUAGGGGGUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGCUUAG-CGUAAG-CGAAGGUAUGAAUUGAAGCCCCAGUAAACGGCGGC .(((((((((...(((....)))...))..)))))))...(((.(((...((((((..((((..((...((((((.(-(....)-)..))))))...))..))))))))))...)))))) ( -43.70) >sp_ag.0 2139123 118 - 2160267/1000-1120 AGCUCUCUGCUAAAUCGUAAGAUGAUGUAUAGGGGGUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGCUUAG-CGUAAG-CGAAGGUAUGAAUUGAAGCCCCAGUAAACGGCGGC .(((((((((...(((....)))...))..)))))))...(((.(((...((((((..((((..((...((((((.(-(....)-)..))))))...))..))))))))))...)))))) ( -43.70) >sp_mu.0 1684213 120 + 2030921/1000-1120 AGCUCUCUGCGAAAUCGUAAGAUGAAGUAUAGGGGGUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGCGCUUAGACGUUUGUCGAAGGUGUGAAUUGAAGCCCCAGUAAACGGCGGC .(((((((((...(((....)))...))..)))))))...(((.(((...((((((..((((..((...((((((.(((....)))..))))))...))..))))))))))...)))))) ( -43.70) >consensus AGCUCUCUGCUAAAUCGUAAGAUGAUGUAUAGGGGGUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGUGCUUAG_CGUAAG_CGAAGGUAUGAAUUGAAGCCCCAGUAAACGGCGGC .(((((((((...(((....)))...))..)))))))...(((.(((...((((((..((((..((...((((((.(((....)))..))))))...))..))))))))))...)))))) (-41.71 = -41.43 + -0.28) # Strand winner: reverse (0.99)

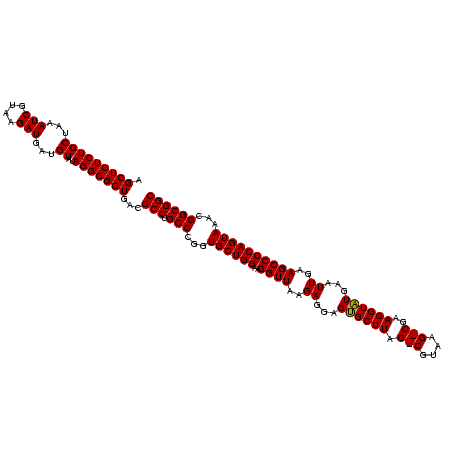
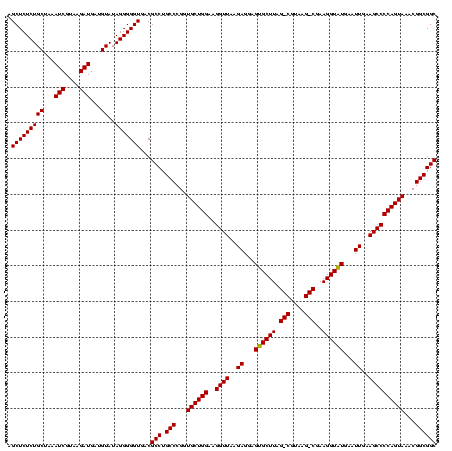
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -39.10 |
| Consensus MFE | -37.35 |
| Energy contribution | -36.80 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.887006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/1080-1200 CCCCGUAACUUCGGGAGAAGGGGUGCUGACUUUAAGUCAGCCGCAGUGAAUAGGCCCAAGCAACUGUUUAUCAAAAACACAGCUCUCUGCUAAAUCGUAAGAUGAUGUAUAGGGGGUGAC ((((((..((((....))))((((((((((.....))))))............))))..))..((((.((((........(((.....)))..(((....))))))).)))))))).... ( -38.00) >sp_ag.0 2139123 120 - 2160267/1080-1200 CCCCGUAACUUCGGGAGAAGGGGCGCUGAUUUUAGAUCAGCCGCAGUGAAUAGGCCCAAGCAACUGUUUAUCAAAAACACAGCUCUCUGCUAAAUCGUAAGAUGAUGUAUAGGGGGUGAC ((((((..((((....))))(((((((((((...)))))))..(........)))))..))..((((.((((........(((.....)))..(((....))))))).)))))))).... ( -38.00) >sp_mu.0 1684213 120 + 2030921/1080-1200 CCCCGUAACUUCGGGAGAAGGGGCGCUGGCGAUAAGUCAGCCGCAGUGAAAAGGCCCAAGCAACUGUUUAUCAAAAACACAGCUCUCUGCGAAAUCGUAAGAUGAAGUAUAGGGGGUGAC ((((.(((((((.(((((..((((((((((.....)))))).....(....).))))..((...(((((.....)))))..))))))).....(((....)))))))).)).)))).... ( -41.30) >consensus CCCCGUAACUUCGGGAGAAGGGGCGCUGACUUUAAGUCAGCCGCAGUGAAUAGGCCCAAGCAACUGUUUAUCAAAAACACAGCUCUCUGCUAAAUCGUAAGAUGAUGUAUAGGGGGUGAC ((((((..((((....))))((((((((((.....))))))..(........)))))..))..((((.((((.........((.....))...(((....))))))).)))))))).... (-37.35 = -36.80 + -0.55) # Strand winner: reverse (0.99)

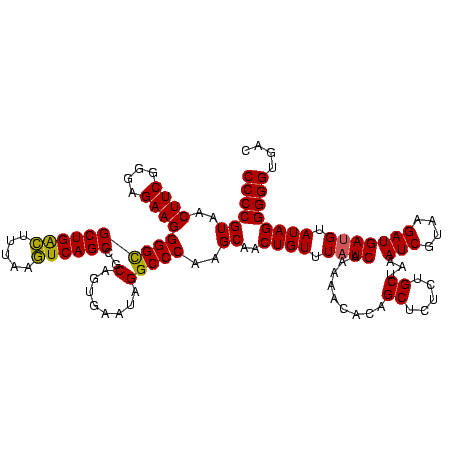
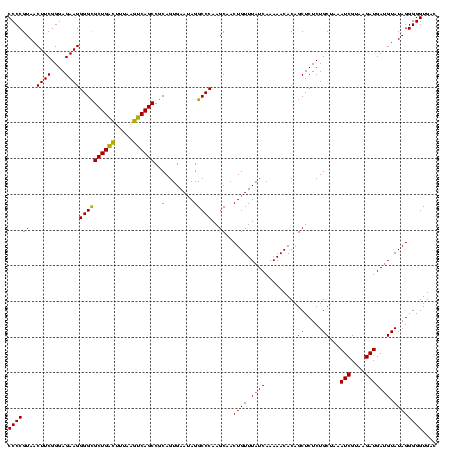
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -37.00 |
| Consensus MFE | -35.31 |
| Energy contribution | -35.20 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.614994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/1120-1240 GUGAGCGAGAGAACUCUCGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUGACUUUAAGUCAGCCGCAGUGAAUAGGCCCAAGCAACUGUUUAUCAAAAACAC ((((((((((....)))))))..........((......(((((....(....).....)))))((((((.....)))))).)).((((((((((....))..))))))))......))) ( -37.30) >sp_ag.0 2139123 120 - 2160267/1120-1240 GUGAGCGAGAGAACUCUCGUUAAGGAACUCGGCAAAAUGGCCCCGUAACUUCGGGAGAAGGGGCGCUGAUUUUAGAUCAGCCGCAGUGAAUAGGCCCAAGCAACUGUUUAUCAAAAACAC ((((((((((....)))))))..........((......(((((....(....).....)))))(((((((...))))))).)).((((((((((....))..))))))))......))) ( -37.80) >sp_mu.0 1684213 120 + 2030921/1120-1240 GUGAUCGAGCGAACUCUCGUUAAGGAACUCGGCAAAAUGGCCCCGUAACUUCGGGAGAAGGGGCGCUGGCGAUAAGUCAGCCGCAGUGAAAAGGCCCAAGCAACUGUUUAUCAAAAACAC .((((.(((((..((((((..(((..((..(((......)))..))..)))))))))...((((((((((.....)))))).....(....).)))).......)))))))))....... ( -35.90) >consensus GUGAGCGAGAGAACUCUCGUUAAGGAACUCGGCAAAAUGGCCCCGUAACUUCGGGAGAAGGGGCGCUGACUUUAAGUCAGCCGCAGUGAAUAGGCCCAAGCAACUGUUUAUCAAAAACAC ((((((((((....)))))))..........((......(((((....(....).....)))))((((((.....)))))).)).((((((((((....))..))))))))......))) (-35.31 = -35.20 + -0.11) # Strand winner: reverse (0.87)

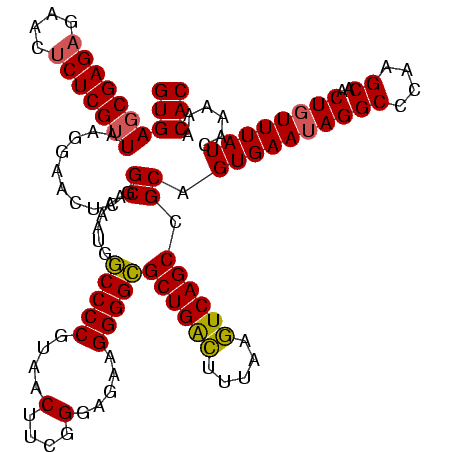
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -41.83 |
| Consensus MFE | -41.64 |
| Energy contribution | -41.20 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.54 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.751822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/1160-1280 ACCGCAAACCGACACAGGUAGUCGAGGCGAGUAGCCUCAGGUGAGCGAGAGAACUCUCGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUGACUUUAAGUCAG .(((...((((((.......)))(((((.....))))).))).(((((((....)))))))........))).......(((((....(....).....))))).(((((.....))))) ( -42.70) >sp_ag.0 2139123 120 - 2160267/1160-1280 ACCGCAAACCGACACAGGUAGUCGAGGCGAGUAGCCUCAGGUGAGCGAGAGAACUCUCGUUAAGGAACUCGGCAAAAUGGCCCCGUAACUUCGGGAGAAGGGGCGCUGAUUUUAGAUCAG .((....((((((.......)))(((((.....))))).))).(((((((....)))))))..))...(((((......(((((....(....).....))))))))))........... ( -43.00) >sp_mu.0 1684213 120 + 2030921/1160-1280 ACCGCAAACCGACACAGGUAGUCGAGGCGAGUAGCCUCAGGUGAUCGAGCGAACUCUCGUUAAGGAACUCGGCAAAAUGGCCCCGUAACUUCGGGAGAAGGGGCGCUGGCGAUAAGUCAG .((....((((((.......)))(((((.....))))).))).....(((((....)))))..)).....(((...((.(((.(((..((((....))))..)))..))).))..))).. ( -39.80) >consensus ACCGCAAACCGACACAGGUAGUCGAGGCGAGUAGCCUCAGGUGAGCGAGAGAACUCUCGUUAAGGAACUCGGCAAAAUGGCCCCGUAACUUCGGGAGAAGGGGCGCUGACUUUAAGUCAG .(((...((((((.......)))(((((.....))))).))).(((((((....)))))))........))).......(((((....(....).....))))).(((((.....))))) (-41.64 = -41.20 + -0.44) # Strand winner: reverse (0.99)

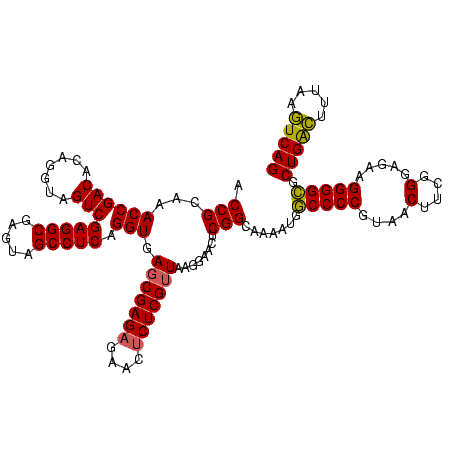
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -40.67 |
| Consensus MFE | -37.70 |
| Energy contribution | -38.03 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.786279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/1200-1320 UCAUUUUGCCGAGUUCCUUAACGAGAGUUCUCUCGCUCACCUGAGGCUACUCGCCUCGACUACCUGUGUCGGUUUGCGGUACGGGUAGAGUAUGGUUAAACGCUAGAAGCUUUUCUUGGC .......((((((....(((((((((....))))((((...((((((.....))))))..(((((((..((.....))..)))))))))))...)))))..((.....))....)))))) ( -42.80) >sp_ag.0 2139123 119 - 2160267/1200-1320 CCAUUUUGCCGAGUUCCUUAACGAGAGUUCUCUCGCUCACCUGAGGCUACUCGCCUCGACUACCUGUGUCGGUUUGCGGUACGGGUAGAGUAUAUGUAU-CGCUAGAAGCUUUUCUUGGC .......((((((.........(((((((.(((.((.....((((((.....)))))).((((((((..((.....))..))))))))...........-.)).)))))))))))))))) ( -40.80) >sp_mu.0 1684213 120 + 2030921/1200-1320 CCAUUUUGCCGAGUUCCUUAACGAGAGUUCGCUCGAUCACCUGAGGCUACUCGCCUCGACUACCUGUGUCGGUUUGCGGUACGGGUAGAGUAUUCAUUAACGCUAGAAGCUUUUCUUGGC .......((((((...(((..((.(((....)))((..((.((((((.....)))))).((((((((..((.....))..)))))))).))..)).....))....))).....)))))) ( -38.40) >consensus CCAUUUUGCCGAGUUCCUUAACGAGAGUUCUCUCGCUCACCUGAGGCUACUCGCCUCGACUACCUGUGUCGGUUUGCGGUACGGGUAGAGUAUACAUAAACGCUAGAAGCUUUUCUUGGC .......((((((........(((((....)))))......((((((.....)))))).((((((((..((.....))..)))))))).............((.....))....)))))) (-37.70 = -38.03 + 0.33)

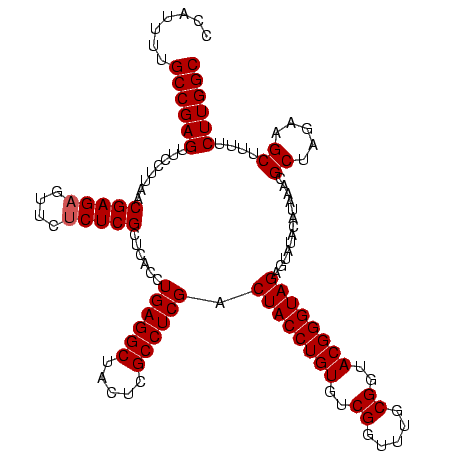
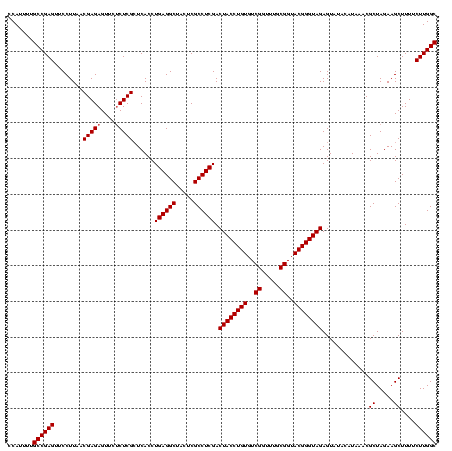
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.06 |
| Mean single sequence MFE | -36.60 |
| Consensus MFE | -37.32 |
| Energy contribution | -33.23 |
| Covariance contribution | -4.09 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.86 |
| Structure conservation index | 1.02 |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.998749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/1320-1440 GCAGACGAUUGGAAGAGUCUGUCUAAGCAGUGAGGUGUGAAUUGAGUCAAAUGCUUAAUUCUAUAACAUUGAGCUGUGAUGGGGAGCGAAGUUUAGUAGCGAAGUUAGUGACGUCACACU ((((((((((.....)))).))))...(((((..(((.(((((((((.....))))))))))))..))))).))(((((((...(((...(((....)))...))).....))))))).. ( -32.40) >sp_ag.0 2139123 120 - 2160267/1320-1440 CCAGACGAUUGGAAGUGUCUGGUCAAACAGUGAGGUGUGAUAUGAGUCAAAUGCUUAUAUCUUUAACAUUGAGCUGUGACGAGGAGCGAAGUUUAGUAGCGAAGUUAGUGAUGUCACACU (((((((........)))))))...........((((((((((.(((((...(((((............)))))..))))....(((...(((....)))...)))..).)))))))))) ( -33.30) >sp_mu.0 1684213 120 + 2030921/1320-1440 GCGUGCGAUUGGAAGAGCACGUCCAAGCAGUGAGGUGAGGACUGAGUCAAAUGCUUAGUUCUGUGCCACCAAGCUGUGACGGGGAGCGAAGUUUAGUAGCGAAGCUAGUGAUGUCACUCU ((((((..........))))))((..(((((..((((((((((((((.....))))))))))....))))..)))))...))((((.((...(((.((((...)))).)))..)).)))) ( -44.10) >consensus GCAGACGAUUGGAAGAGUCUGUCCAAGCAGUGAGGUGUGAAAUGAGUCAAAUGCUUAAUUCUAUAACAUUGAGCUGUGACGGGGAGCGAAGUUUAGUAGCGAAGUUAGUGAUGUCACACU ((((((..........))))))....(((((..((((((((((((((.....)))))))))....)))))..)))))((((...(((...(((....)))...))).....))))..... (-37.32 = -33.23 + -4.09) # Strand winner: reverse (1.00)

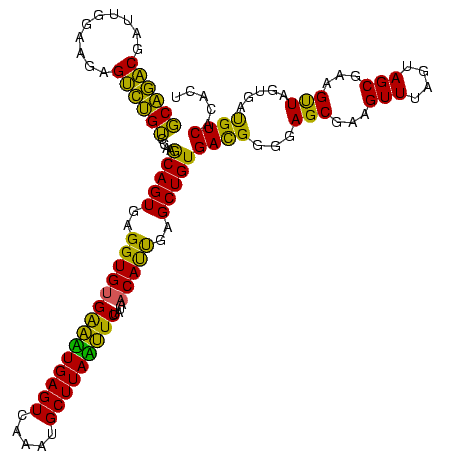
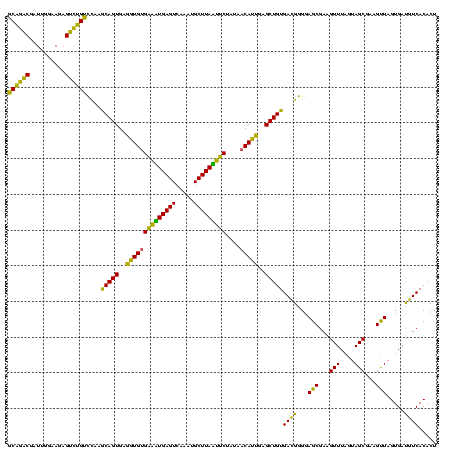
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.22 |
| Mean single sequence MFE | -34.03 |
| Consensus MFE | -31.53 |
| Energy contribution | -28.10 |
| Covariance contribution | -3.43 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.981834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/1360-1480 AGAGUAUGUAGUGAUGGAGGGACGCAGUAGGCUAACUAAAGCAGACGAUUGGAAGAGUCUGUCUAAGCAGUGAGGUGUGAAUUGAGUCAAAUGCUUAAUUCUAUAACAUUGAGCUGUGAU ..............((((.((((((..(((.....)))..))...(........).)))).)))).(((((..((((((((((((((.....)))))))))....)))))..)))))... ( -31.20) >sp_ag.0 2139123 120 - 2160267/1360-1480 AGAGUAUAUAGUGAUGGAGGGACGCAGUAGGCUAACUAAACCAGACGAUUGGAAGUGUCUGGUCAAACAGUGAGGUGUGAUAUGAGUCAAAUGCUUAUAUCUUUAACAUUGAGCUGUGAC ..(((...((.((.((......)))).)).)))......((((((((........))))))))...(((((..((((((((((((((.....)))))))))....)))))..)))))... ( -32.50) >sp_mu.0 1684213 120 + 2030921/1360-1480 AGAGUAUUGAGUGAAGGAGGGACGCAGCAGGCUAACUAGAGCGUGCGAUUGGAAGAGCACGUCCAAGCAGUGAGGUGAGGACUGAGUCAAAUGCUUAGUUCUGUGCCACCAAGCUGUGAC ...................(((((..(((.(((......))).)))...((......)))))))..(((((..((((((((((((((.....))))))))))....))))..)))))... ( -38.40) >consensus AGAGUAUAUAGUGAUGGAGGGACGCAGUAGGCUAACUAAAGCAGACGAUUGGAAGAGUCUGUCCAAGCAGUGAGGUGUGAAAUGAGUCAAAUGCUUAAUUCUAUAACAUUGAGCUGUGAC ..(((.....(((.........))).....))).......((((((..........))))))....(((((..((((((((((((((.....)))))))))....)))))..)))))... (-31.53 = -28.10 + -3.43) # Strand winner: reverse (1.00)

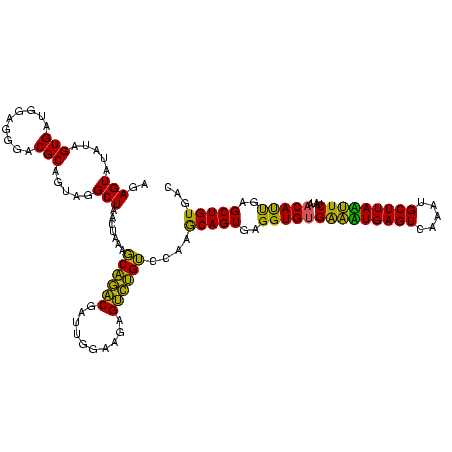
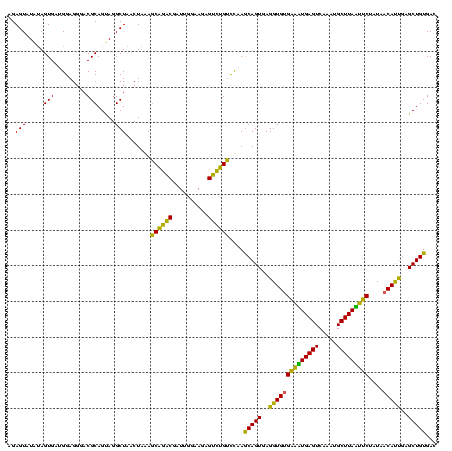
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -21.10 |
| Consensus MFE | -19.56 |
| Energy contribution | -18.23 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/1400-1520 UCACACCUCACUGCUUAGACAGACUCUUCCAAUCGUCUGCUUUAGUUAGCCUACUGCGUCCCUCCAUCACUACAUACUCUAGUACAGGAAUAUCAACCUGUUGUCCAUCGGAUACACCUU ......((.((((...((.(((((..........))))))).)))).)).....((..(((.......((((.......))))(((((........)))))........)))..)).... ( -18.70) >sp_ag.0 2139123 120 - 2160267/1400-1520 UCACACCUCACUGUUUGACCAGACACUUCCAAUCGUCUGGUUUAGUUAGCCUACUGCGUCCCUCCAUCACUAUAUACUCUAGUACAGGAAUAUCAACCUGUUGUCCAUCGGAUACACCUU ......((.((((...((((((((..........)))))))))))).)).....((..(((.......((((.......))))(((((........)))))........)))..)).... ( -23.20) >sp_mu.0 1684213 120 + 2030921/1400-1520 CCUCACCUCACUGCUUGGACGUGCUCUUCCAAUCGCACGCUCUAGUUAGCCUGCUGCGUCCCUCCUUCACUCAAUACUCUAGUACAGGAAUAUCAACCUGUUGCCCAUCGGAUACACCUA .....((.........((((((((..........))))((..(((.....)))..))))))....................(((((((........)))).))).....))......... ( -21.40) >consensus UCACACCUCACUGCUUGGACAGACUCUUCCAAUCGUCUGCUUUAGUUAGCCUACUGCGUCCCUCCAUCACUAAAUACUCUAGUACAGGAAUAUCAACCUGUUGUCCAUCGGAUACACCUU ..........(((.((((((((((..........))))).))))).))).....((..(((.......((((.......))))(((((........)))))........)))..)).... (-19.56 = -18.23 + -1.33)

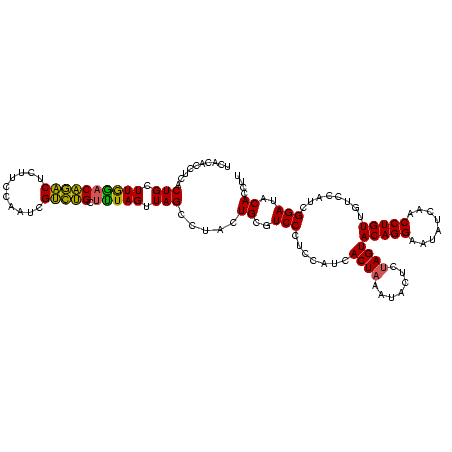
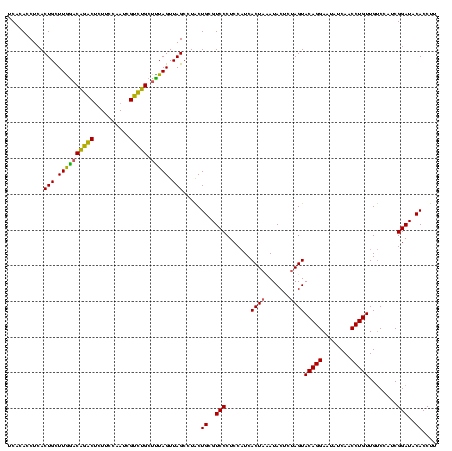
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -40.23 |
| Consensus MFE | -39.30 |
| Energy contribution | -39.63 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917763 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/1480-1600 AGUACAGGAAUAUCAACCUGUUGUCCAUCGGAUACACCUUUCGGUCUCUCCUUAGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAACCUUAGUCUUACGGUGGACAGGAUU ...(((((........)))))(((((((((.....((((...((.....))..))))...((((((.((((((.(((......))))))))).....))))))...)))))))))..... ( -41.40) >sp_ag.0 2139123 120 - 2160267/1480-1600 AGUACAGGAAUAUCAACCUGUUGUCCAUCGGAUACACCUUUCGGUCUCUCCUUAGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAACCUUAGUCUUACGGUGGACAGGAUU ...(((((........)))))(((((((((.....((((...((.....))..))))...((((((.((((((.(((......))))))))).....))))))...)))))))))..... ( -41.40) >sp_mu.0 1684213 120 + 2030921/1480-1600 AGUACAGGAAUAUCAACCUGUUGCCCAUCGGAUACACCUAUCGGUCUCUCCUUAGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAUCCUUAGUCUUACGGUGGACAGGAUU ................(((((...((((((.....(((((..((.....)).)))))...((((((.((((((.(((......))))))))).....))))))...)))))))))))... ( -37.90) >consensus AGUACAGGAAUAUCAACCUGUUGUCCAUCGGAUACACCUUUCGGUCUCUCCUUAGGUCCCGACUAACCCAGGGCGGACGAGCCUUCCCCUGGAAACCUUAGUCUUACGGUGGACAGGAUU ...(((((........)))))(((((((((.....((((...((.....))..))))...((((((.((((((.(((......))))))))).....))))))...)))))))))..... (-39.30 = -39.63 + 0.33)

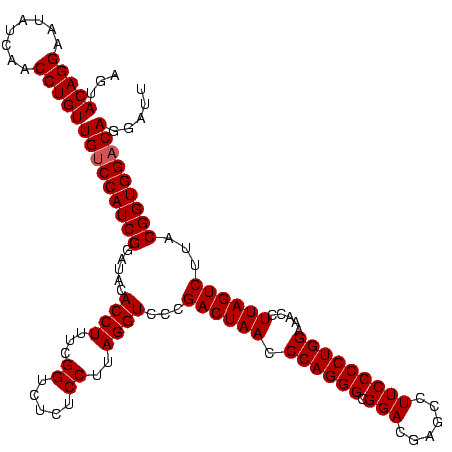
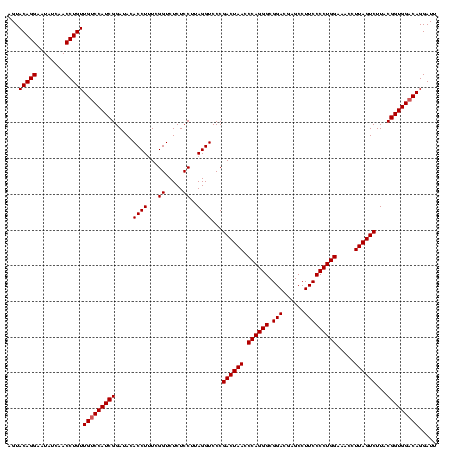
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -44.10 |
| Consensus MFE | -44.32 |
| Energy contribution | -44.10 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.04 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/1480-1600 AAUCCUGUCCACCGUAAGACUAAGGUUUCCAGGGGAAGGCUCGUCCGCCCUGGGUUAGUCGGGACCUAAGGAGAGACCGAAAGGUGUAUCCGAUGGACAACAGGUUGAUAUUCCUGUACU .....((((((.(((..((((((....(((((((...((.....)).)))))))))))))...))....(((.(.(((....))).).)))).))))))(((((........)))))... ( -45.30) >sp_ag.0 2139123 120 - 2160267/1480-1600 AAUCCUGUCCACCGUAAGACUAAGGUUUCCAGGGGAAGGCUCGUCCGCCCUGGGUUAGUCGGGACCUAAGGAGAGACCGAAAGGUGUAUCCGAUGGACAACAGGUUGAUAUUCCUGUACU .....((((((.(((..((((((....(((((((...((.....)).)))))))))))))...))....(((.(.(((....))).).)))).))))))(((((........)))))... ( -45.30) >sp_mu.0 1684213 120 + 2030921/1480-1600 AAUCCUGUCCACCGUAAGACUAAGGAUUCCAGGGGAAGGCUCGUCCGCCCUGGGUUAGUCGGGACCUAAGGAGAGACCGAUAGGUGUAUCCGAUGGGCAACAGGUUGAUAUUCCUGUACU .....((((((.(((..((((((....(((((((...((.....)).)))))))))))))...))....(((.(.(((....))).).)))).))))))(((((........)))))... ( -41.70) >consensus AAUCCUGUCCACCGUAAGACUAAGGUUUCCAGGGGAAGGCUCGUCCGCCCUGGGUUAGUCGGGACCUAAGGAGAGACCGAAAGGUGUAUCCGAUGGACAACAGGUUGAUAUUCCUGUACU .....((((((.(((..((((((....(((((((...((.....)).)))))))))))))...))....(((.(.(((....))).).)))).))))))(((((........)))))... (-44.32 = -44.10 + -0.22) # Strand winner: reverse (0.98)

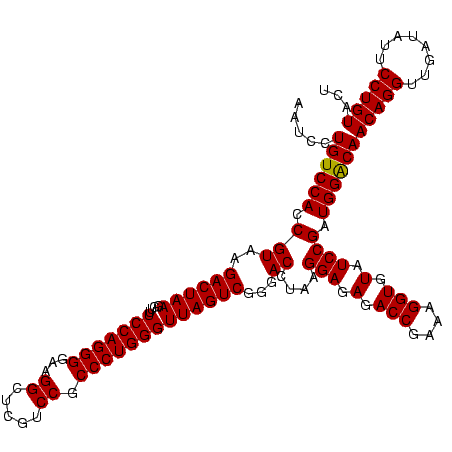
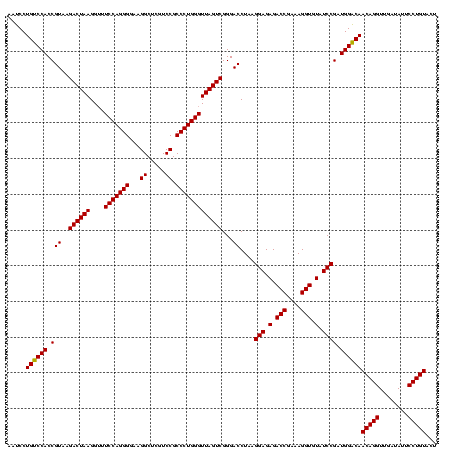
| Location | 1,790,020 – 1,790,138 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.75 |
| Mean single sequence MFE | -37.84 |
| Consensus MFE | -35.75 |
| Energy contribution | -33.76 |
| Covariance contribution | -1.99 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 118 + 2038615/1640-1760 CGCCGAAAAUGUACCGGGGCUAAAACAAUUUACCGAAGCUGUGGAUACCUUUAUA--GGUAUGGUAGGAGAGCGUUCUAUGUGUGAUGAAGGUAUACCGUGAGGAGUGCUGGAACGCAUA .(((.........(((.((((...............)))).)))((((((....)--))))))))......(((((((............(((((.((....)).))))))))))))... ( -35.06) >sp_ag.0 2139123 120 - 2160267/1640-1760 CGCCGAAAAUGUACCGGGGCUAAAACAUAUUACCGAAGCUGUGGAUGACACAAAAGUGUCAUGGUAGGAGAGCGUUCUAUGUGUGAAGAAGGUGUACCGUGAGGAGCGCUGGAACGCAUA .(((.........(((.((((...............)))).)))(((((((....))))))))))......(((((((............(((((.((....)).))))))))))))... ( -39.36) >sp_mu.0 1684213 116 + 2030921/1640-1760 CGCCGAAAAUGUACCGGGGCUGAAACAAUUUACCGAAGCUGUGGAUCCCUUAG----GGGAUGGUAGGAGAGCGUUCUAUGUGCGCAGAAGGUGUACCGCAAGGAGCGCUGGAGUGCAUA ........(((((((((.(((.........((((....(((((.(((((....----))))).((((((.....))))))...)))))..))))..((....))))).))))..))))). ( -39.10) >consensus CGCCGAAAAUGUACCGGGGCUAAAACAAUUUACCGAAGCUGUGGAUACCUUAA_A__GGCAUGGUAGGAGAGCGUUCUAUGUGUGAAGAAGGUGUACCGUGAGGAGCGCUGGAACGCAUA .(((.........(((.((((...............)))).)))(((((........))))))))......(((((((............(((((.((....)).))))))))))))... (-35.75 = -33.76 + -1.99) # Strand winner: reverse (1.00)

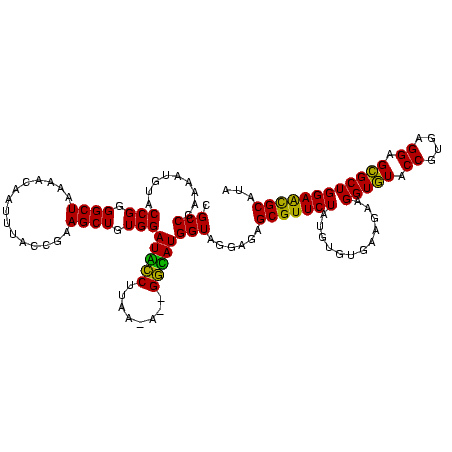
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -27.29 |
| Consensus MFE | -26.43 |
| Energy contribution | -26.43 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.656119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/1760-1880 CAGGGUCACUCGACUAGUGAGCUAUUACGCACUCUUUGAAUGAAUAGCUGCUUCUAAGCUAACAUCCUAGUUGUCUGUGCAACCCCACAUCCUUUUCCACUUAACAAUUAUUUUGGGACC ..(((((((.(((((((.((((((((...((.....))....)))))).(((....))).....)))))))))...)))..)))).........(((((..............))))).. ( -26.64) >sp_ag.0 2139123 120 - 2160267/1760-1880 CAGGGUCACUCGACUAGUGAGCUAUUACGCACUCUUUGAAUGAAUAGCUGCUUCUAAGCUAACAUCCUAGUUGUCUGUGCAACCCCACAUCCUUUUCCACUUAACAUAUAUUUUGGGACC ..(((((((.(((((((.((((((((...((.....))....)))))).(((....))).....)))))))))...)))..)))).........(((((..............))))).. ( -26.64) >sp_mu.0 1684213 120 + 2030921/1760-1880 CAGGGUCACUCGACUAGUGAGCUAUUACGCACUCUUUGAAUGAAUAGCUGCUUCUAAGCUAACAUCCUAGUUGUCUGUGCAACCCCACAUCCUUUUCCACUUAACAAUUAUUUGGGGACC ..(((((((.(((((((.((((((((...((.....))....)))))).(((....))).....)))))))))...)))..))))........(..(((..((.....))..)))..).. ( -28.60) >consensus CAGGGUCACUCGACUAGUGAGCUAUUACGCACUCUUUGAAUGAAUAGCUGCUUCUAAGCUAACAUCCUAGUUGUCUGUGCAACCCCACAUCCUUUUCCACUUAACAAUUAUUUUGGGACC ..(((((((.(((((((.((((((((...((.....))....)))))).(((....))).....)))))))))...)))..))))....((((.....................)))).. (-26.43 = -26.43 + 0.00)

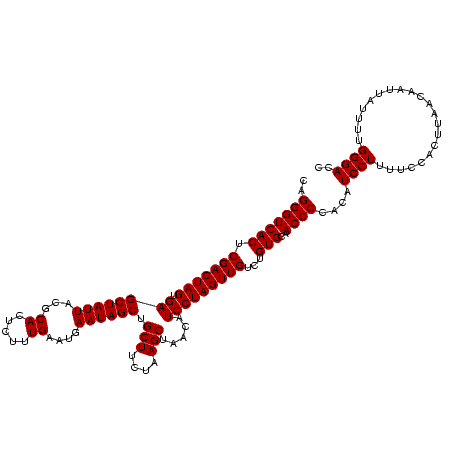
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -32.93 |
| Consensus MFE | -33.82 |
| Energy contribution | -32.93 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.65 |
| Structure conservation index | 1.03 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991543 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/2080-2200 UCUCCAAGUUCGUUUGGAAUUUCUCCGCUACCCACAAGUCAUCCAAGCACUUUUCAACGUGCCCUGGUUCGGUCCUCCAGUGCGUCUUACCGCACCUUCAACCUGCUCAUGGGUAGGUCA ..((((((....)))))).........(((((((..(((...(((.((((........))))..)))...(((......(((((......))))).....))).)))..))))))).... ( -34.00) >sp_ag.0 2139123 120 - 2160267/2080-2200 UCUCCAAGUUCGUUUGGAAUUUCUCCGCUACCCACAAGUCAUCCAAGCACUUUUCAACGUGCCCUGGUUCGGUCCUCCAGUGAGUUUUACCUCACCUUCAACCUGCUCAUGGGUAGGUCA ..((((((....)))))).........(((((((..(((...(((.((((........))))..)))...(((......(((((......))))).....))).)))..))))))).... ( -32.70) >sp_mu.0 1684213 120 + 2030921/2080-2200 UCUCCAAGUUCGUUUGGAAUUUCUCCGCUACCCACAAGUCAUCCAAACACUUUUCAACGUGUCCUGGUUCGGUCCUCCAGUGCGUUUUACCGCACCUUCAACCUGCUCAUGGGUAGGUCA ..((((((....)))))).........(((((((..(((...(((.((((........))))..)))...(((......(((((......))))).....))).)))..))))))).... ( -32.10) >consensus UCUCCAAGUUCGUUUGGAAUUUCUCCGCUACCCACAAGUCAUCCAAGCACUUUUCAACGUGCCCUGGUUCGGUCCUCCAGUGCGUUUUACCGCACCUUCAACCUGCUCAUGGGUAGGUCA ..((((((....)))))).........(((((((..(((...(((.((((........))))..)))...(((......(((((......))))).....))).)))..))))))).... (-33.82 = -32.93 + -0.89)

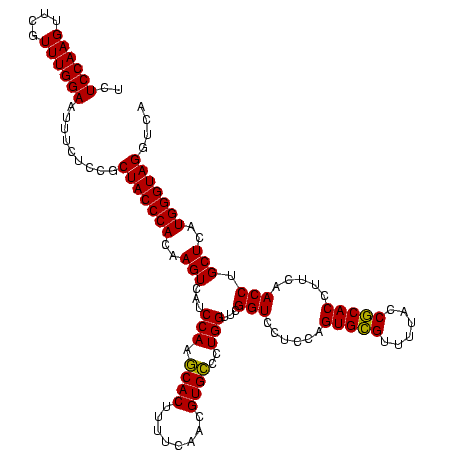
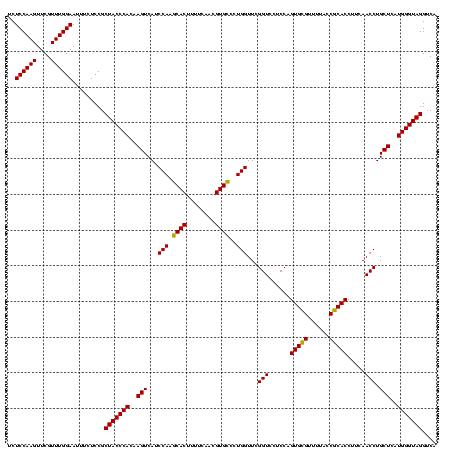
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -46.90 |
| Consensus MFE | -47.79 |
| Energy contribution | -46.90 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.51 |
| Structure conservation index | 1.02 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/2080-2200 UGACCUACCCAUGAGCAGGUUGAAGGUGCGGUAAGACGCACUGGAGGACCGAACCAGGGCACGUUGAAAAGUGCUUGGAUGACUUGUGGGUAGCGGAGAAAUUCCAAACGAACUUGGAGA ...((((((((..((((((((...((((((......))))))....))))...(((((.(((........)))))))).)).))..))))))).)......((((((......)))))). ( -48.00) >sp_ag.0 2139123 120 - 2160267/2080-2200 UGACCUACCCAUGAGCAGGUUGAAGGUGAGGUAAAACUCACUGGAGGACCGAACCAGGGCACGUUGAAAAGUGCUUGGAUGACUUGUGGGUAGCGGAGAAAUUCCAAACGAACUUGGAGA ...((((((((..((((((((...((((((......))))))....))))...(((((.(((........)))))))).)).))..))))))).)......((((((......)))))). ( -46.70) >sp_mu.0 1684213 120 + 2030921/2080-2200 UGACCUACCCAUGAGCAGGUUGAAGGUGCGGUAAAACGCACUGGAGGACCGAACCAGGACACGUUGAAAAGUGUUUGGAUGACUUGUGGGUAGCGGAGAAAUUCCAAACGAACUUGGAGA ...((((((((..((((((((...((((((......))))))....))))...((((.((((........)))))))).)).))..))))))).)......((((((......)))))). ( -46.00) >consensus UGACCUACCCAUGAGCAGGUUGAAGGUGCGGUAAAACGCACUGGAGGACCGAACCAGGGCACGUUGAAAAGUGCUUGGAUGACUUGUGGGUAGCGGAGAAAUUCCAAACGAACUUGGAGA ...((((((((..((((((((...((((((......))))))....))))...((((.((((........)))))))).)).))..))))))).)......((((((......)))))). (-47.79 = -46.90 + -0.89) # Strand winner: reverse (1.00)

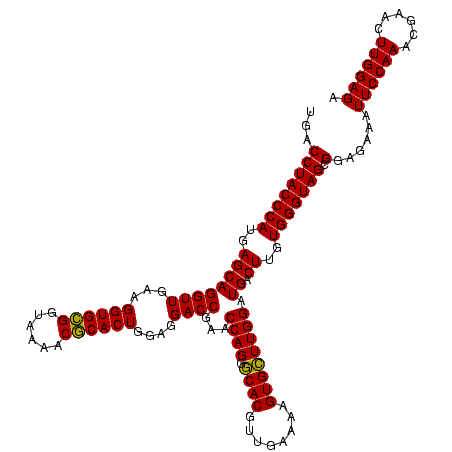
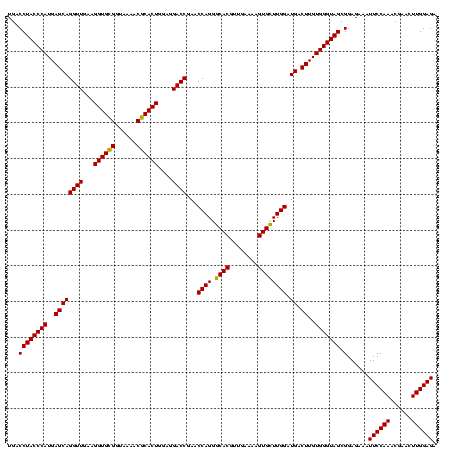
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.72 |
| Mean single sequence MFE | -32.81 |
| Consensus MFE | -30.49 |
| Energy contribution | -29.60 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699143 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/2120-2240 AUCCAAGCACUUUUCAACGUGCCCUGGUUCGGUCCUCCAGUGCGUCUUACCGCACCUUCAACCUGCUCAUGGGUAGGUCACAUGGUUUCGGGUCUACGUCAUGAUACUAAGGUGCCCUAU ......(((((((....(((((...((..(((....((((((((......))))).....(((((((....)))))))....)))..)))..))...).)))).....)))))))..... ( -36.10) >sp_ag.0 2139123 120 - 2160267/2120-2240 AUCCAAGCACUUUUCAACGUGCCCUGGUUCGGUCCUCCAGUGAGUUUUACCUCACCUUCAACCUGCUCAUGGGUAGGUCACAUGGUUUCGGGUCUACAACAUGAUACUAUGACGCCCUAU ..(((.((((........))))..)))...(((......(((((......))))).....(((((((....)))))))..((((((.(((.((.....)).))).))))))..))).... ( -31.60) >sp_mu.0 1684213 120 + 2030921/2120-2240 AUCCAAACACUUUUCAACGUGUCCUGGUUCGGUCCUCCAGUGCGUUUUACCGCACCUUCAACCUGCUCAUGGGUAGGUCACUUGGUUUCGGGUCUACAUCUACGUACUUAACCGCCCUUU ..(((.((((........))))..)))..((((.....(((((((....(((.(((....(((((((....))))))).....)))..)))..........)))))))..))))...... ( -30.74) >consensus AUCCAAGCACUUUUCAACGUGCCCUGGUUCGGUCCUCCAGUGCGUUUUACCGCACCUUCAACCUGCUCAUGGGUAGGUCACAUGGUUUCGGGUCUACAUCAUGAUACUAAGACGCCCUAU ..(((.((((........))))..)))...(..((.((((((((......))))).....(((((((....)))))))....)))....))..).......................... (-30.49 = -29.60 + -0.89)

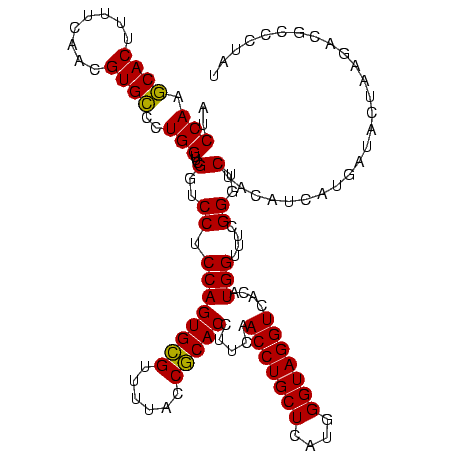
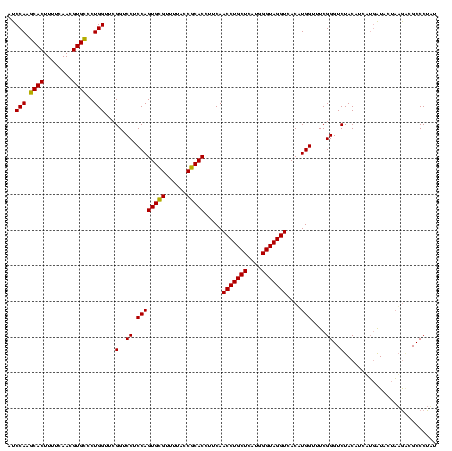
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.17 |
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -39.30 |
| Energy contribution | -36.87 |
| Covariance contribution | -2.44 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.39 |
| Structure conservation index | 1.05 |
| SVM decision value | 3.08 |
| SVM RNA-class probability | 0.998355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/2200-2320 UUUUGUAGAAUGAACCGGCGAGUUACGUUAUGAUGCGAGGUUAAGUUGAAGAGACGGAGCCGUAGGGAAACCGAGUCUGAAUAGGGCACCUUAGUAUCAUGACGUAGACCCGAAACCAUG ...............(((....(((((((((((((((((((............(((....))).((....))..((((.....))))))))).))))))))))))))..)))........ ( -37.80) >sp_ag.0 2139123 120 - 2160267/2200-2320 UUUUGUAGAAUGAACCGGCGAGUUACGAUAUGAUGCGAGGUUAAGUUGAAGAGACGGAGCCGUAGGGAAACCGAGUCUUAAUAGGGCGUCAUAGUAUCAUGUUGUAGACCCGAAACCAUG ...............(((....((((((((((((((..((((..(((.....)))..))))...((....))((((((.....)))).))...))))))))))))))..)))........ ( -36.60) >sp_mu.0 1684213 120 + 2030921/2200-2320 UUUUGUAGAAUGAACCGGCGAGUUACGUUUACGUGCGAGGUUAAGUUGAAGAGACGGAGCCGUAGGGAAACCGAGUCUGAAAAGGGCGGUUAAGUACGUAGAUGUAGACCCGAAACCAAG ...............(((....((((((((((((((..((((..(((.....)))..)))).......(((((..(((....))).)))))..))))))))))))))..)))........ ( -37.80) >consensus UUUUGUAGAAUGAACCGGCGAGUUACGUUAUGAUGCGAGGUUAAGUUGAAGAGACGGAGCCGUAGGGAAACCGAGUCUGAAUAGGGCGCCUUAGUAUCAUGAUGUAGACCCGAAACCAUG ...............(((....((((((((((((((..((((..(((.....)))..))))...((....))..((((.....))))......))))))))))))))..)))........ (-39.30 = -36.87 + -2.44) # Strand winner: reverse (1.00)

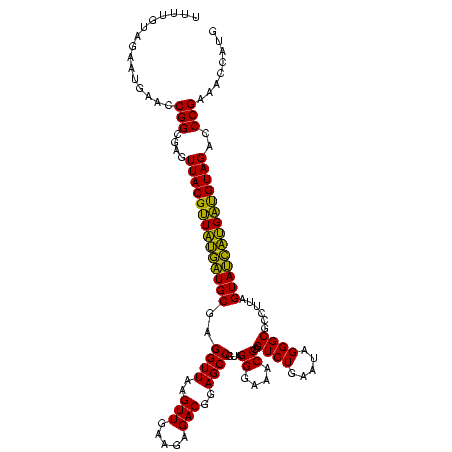

| Location | 1,790,020 – 1,790,139 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -22.34 |
| Energy contribution | -20.23 |
| Covariance contribution | -2.11 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 119 + 2038615/2480-2600 UUCCGACGGGAUUUCACGUGUCCCGCCGUACUCAGGAUACUGCUAAGUACAAAG-ACUAUUUUAAAUACGAGGCUAUUACUCUCUUUGGCUGAUCUUCCCAAAUCAUUCUUCUAUAAUCU .......(((((.......)))))(((((((((((....)))...)))))....-..............(((.......))).....)))((((........)))).............. ( -22.60) >sp_ag.0 2139123 120 - 2160267/2480-2600 UUCCGACGAGAUUUCGCGUGUCUCGCCGUACUCAGGAUACUGCUAAGGUUAAUCUAUCAUUUUAAAUACGAGGCUGUUACUCUCUUUGGCUUACCUUCCCAGGUAAUUCUUCUAUAAUGA ..((((.((((....(((.((((((.......(((....)))....(((......)))..........)))))))))...)))).)))).((((((....)))))).............. ( -23.80) >sp_mu.0 1684213 119 + 2030921/2480-2600 UUCCGACGGGAUUCCUCGUGUCCCGCCGUACUCAGGAUCCUGCUUGGCAUCAAGUGA-AUUUCAAAUACGAGGCUCUUACUCUCUCUGGCGACUCUUCCCAGAGUCUUCUUCUAUUCCCU .......((((..((((((((...((((....(((....)))..)))).((....))-.......))))))))..........((((((.((....))))))))...........)))). ( -33.80) >consensus UUCCGACGGGAUUUCACGUGUCCCGCCGUACUCAGGAUACUGCUAAGCAUAAAGUAA_AUUUUAAAUACGAGGCUAUUACUCUCUUUGGCUAAUCUUCCCAGAUAAUUCUUCUAUAAUCU .......(((((.......)))))(((.....(((....)))...........................(((.......))).....)))((((((....)))))).............. (-22.34 = -20.23 + -2.11)

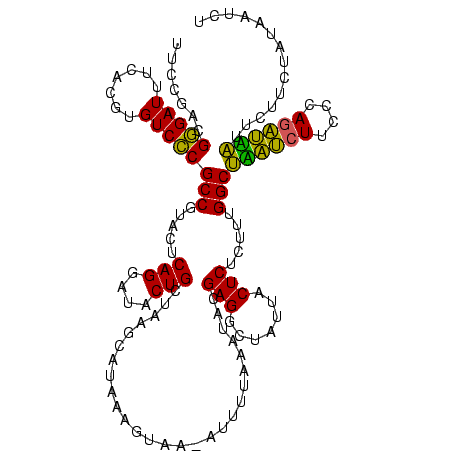
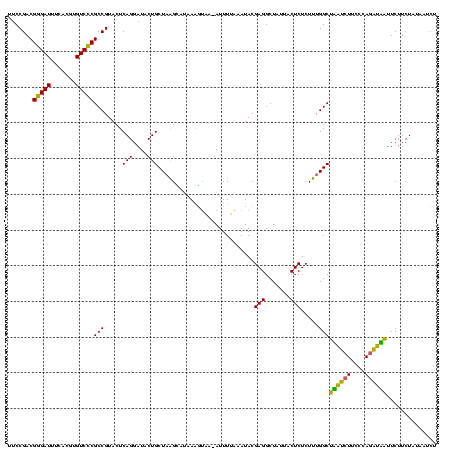
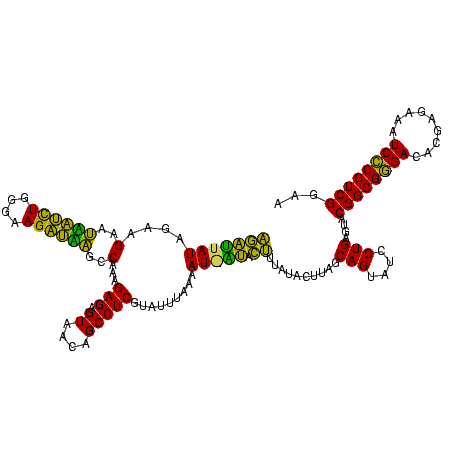
| Location | 1,790,020 – 1,790,139 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.17 |
| Mean single sequence MFE | -31.37 |
| Consensus MFE | -28.82 |
| Energy contribution | -24.50 |
| Covariance contribution | -4.32 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 119 + 2038615/2480-2600 AGAUUAUAGAAGAAUGAUUUGGGAAGAUCAGCCAAAGAGAGUAAUAGCCUCGUAUUUAAAAUAGU-CUUUGUACUUAGCAGUAUCCUGAGUACGGCGGGACACGUGAAAUCCCGUCGGAA (((((((....(..((((((....))))))..)...(((.((....))))).........)))))-))..((((((((.......))))))))(((((((.........))))))).... ( -32.10) >sp_ag.0 2139123 120 - 2160267/2480-2600 UCAUUAUAGAAGAAUUACCUGGGAAGGUAAGCCAAAGAGAGUAACAGCCUCGUAUUUAAAAUGAUAGAUUAACCUUAGCAGUAUCCUGAGUACGGCGAGACACGCGAAAUCUCGUCGGAA (((((.((((.(..((((((....))))))..)...(((.((....)))))...)))).))))).........(((((.......)))))..((((((((.........))))))))... ( -27.70) >sp_mu.0 1684213 119 + 2030921/2480-2600 AGGGAAUAGAAGAAGACUCUGGGAAGAGUCGCCAGAGAGAGUAAGAGCCUCGUAUUUGAAAU-UCACUUGAUGCCAAGCAGGAUCCUGAGUACGGCGGGACACGAGGAAUCCCGUCGGAA .((((.........((((((....))))))(((...(((.((....)))))(((((((..((-((.((((....))))..))))..)))))))))).............))))....... ( -34.30) >consensus AGAUUAUAGAAGAAUAAUCUGGGAAGAUAAGCCAAAGAGAGUAACAGCCUCGUAUUUAAAAU_AUACUUUAUACUUAGCAGUAUCCUGAGUACGGCGGGACACGAGAAAUCCCGUCGGAA (((((((....(..((((((....))))))..)...(((.((....))))).........))))).))..........(((....)))....((((((((.........))))))))... (-28.82 = -24.50 + -4.32) # Strand winner: reverse (1.00)

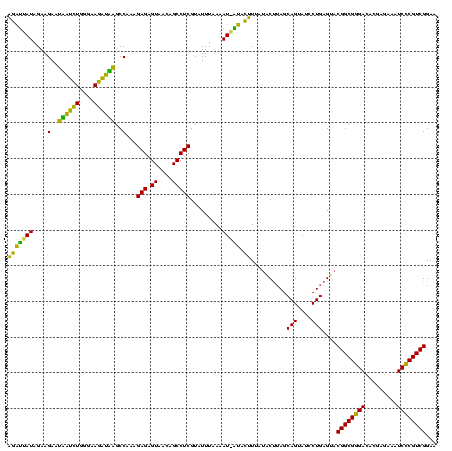
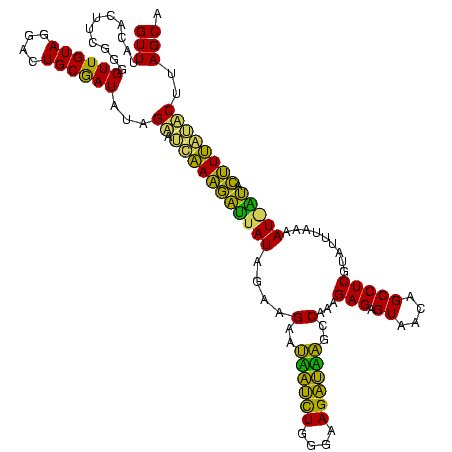
| Location | 1,790,020 – 1,790,139 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.28 |
| Mean single sequence MFE | -29.03 |
| Consensus MFE | -23.27 |
| Energy contribution | -19.07 |
| Covariance contribution | -4.20 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 119 + 2038615/2520-2640 GUUUACUCUUCGGGGUUGUAGGACUGCAAUGUGGACUCAAAGAUUAUAGAAGAAUGAUUUGGGAAGAUCAGCCAAAGAGAGUAAUAGCCUCGUAUUUAAAAUAGU-CUUUGUACUUAGCA ((((((((....))((((((....)))))))))))).((((((((((....(..((((((....))))))..)...(((.((....))))).........)))))-)))))......... ( -27.50) >sp_ag.0 2139123 120 - 2160267/2520-2640 GUUUACACUUCGGGGUUGUAGGACUGCGAUAUAGGAUUAAUCAUUAUAGAAGAAUUACCUGGGAAGGUAAGCCAAAGAGAGUAACAGCCUCGUAUUUAAAAUGAUAGAUUAACCUUAGCA .........(((.((((....)))).)))...(((.(((((((((((....(..((((((....))))))..)...(((.((....))))).........))))).)))))))))..... ( -25.50) >sp_mu.0 1684213 119 + 2030921/2520-2640 GUUUACACUCUGGGGUUGUAGGACUGCGAUAAAGCAGCCAAGGGAAUAGAAGAAGACUCUGGGAAGAGUCGCCAGAGAGAGUAAGAGCCUCGUAUUUGAAAU-UCACUUGAUGCCAAGCA .(((((.((((..(((....((.((((......))))))...............((((((....)))))))))..)))).))))).((...(((((((....-.))...)))))...)). ( -34.10) >consensus GUUUACACUUCGGGGUUGUAGGACUGCGAUAUAGAAUCAAAGAUUAUAGAAGAAUAAUCUGGGAAGAUAAGCCAAAGAGAGUAACAGCCUCGUAUUUAAAAU_AUACUUUAUACUUAGCA (((...........((((((....))))))...((.(((((((((((....(..((((((....))))))..)...(((.((....))))).........))))).))))))))..))). (-23.27 = -19.07 + -4.20) # Strand winner: reverse (1.00)

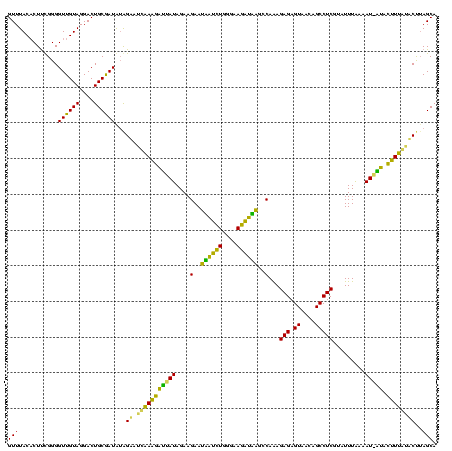
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.83 |
| Mean single sequence MFE | -30.53 |
| Consensus MFE | -30.63 |
| Energy contribution | -27.63 |
| Covariance contribution | -2.99 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.82 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.22 |
| SVM RNA-class probability | 0.998768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/2560-2680 UCUCUUUGGCUGAUCUUCCCAAAUCAUUCUUCUAUAAUCUUUGAGUCCACAUUGCAGUCCUACAACCCCGAAGAGUAAACUCUUCGGUUUGCCCUCCUGCCGUUUCGCUCGCCGCUACUA .......(((((((........))))................((((....((.((((.....(((..((((((((....)))))))).))).....)))).))...)))))))....... ( -27.60) >sp_ag.0 2139123 120 - 2160267/2560-2680 UCUCUUUGGCUUACCUUCCCAGGUAAUUCUUCUAUAAUGAUUAAUCCUAUAUCGCAGUCCUACAACCCCGAAGUGUAAACACUUCGGUUUGCCCUCCUGCCGUUUCGCUCGCCGCUACUA .......(((((((((....))))))...........................((((.....(((..((((((((....)))))))).))).....))))..........)))....... ( -27.70) >sp_mu.0 1684213 120 + 2030921/2560-2680 UCUCUCUGGCGACUCUUCCCAGAGUCUUCUUCUAUUCCCUUGGCUGCUUUAUCGCAGUCCUACAACCCCAGAGUGUAAACACUCUGGUUUGCCCUCCUGCCUCUUCGCUCGCCGCUACUG ......((((((((((....))))))...............(((.((......((((.....(((..((((((((....)))))))).))).....))))......))..)))))))... ( -36.30) >consensus UCUCUUUGGCUAAUCUUCCCAGAUAAUUCUUCUAUAAUCUUUGAUCCUAUAUCGCAGUCCUACAACCCCGAAGUGUAAACACUUCGGUUUGCCCUCCUGCCGUUUCGCUCGCCGCUACUA .......(((((((((....))))))...........................((((.....(((..((((((((....)))))))).))).....))))..........)))....... (-30.63 = -27.63 + -2.99)

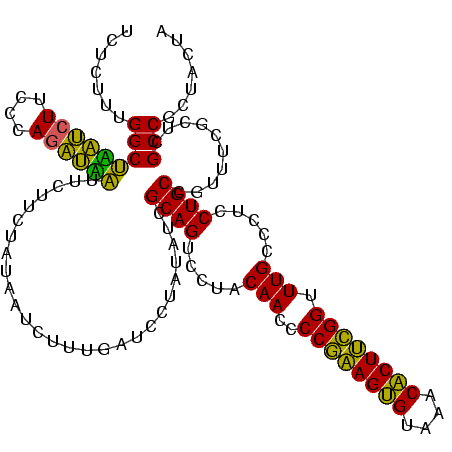
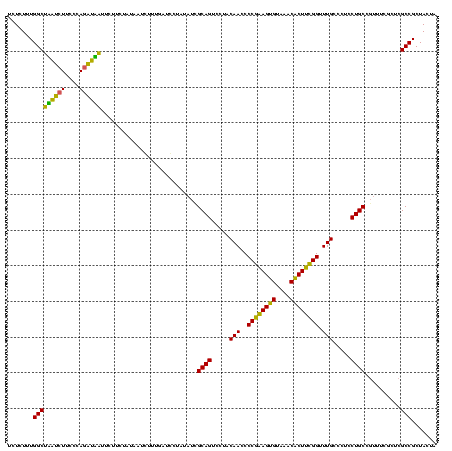
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.83 |
| Mean single sequence MFE | -39.93 |
| Consensus MFE | -40.78 |
| Energy contribution | -37.23 |
| Covariance contribution | -3.55 |
| Combinations/Pair | 1.36 |
| Mean z-score | -3.97 |
| Structure conservation index | 1.02 |
| SVM decision value | 3.12 |
| SVM RNA-class probability | 0.998487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/2560-2680 UAGUAGCGGCGAGCGAAACGGCAGGAGGGCAAACCGAAGAGUUUACUCUUCGGGGUUGUAGGACUGCAAUGUGGACUCAAAGAUUAUAGAAGAAUGAUUUGGGAAGAUCAGCCAAAGAGA .......(((((((.(....((((....((((.((((((((....))))))))..))))....))))....).).)))................((((((....)))))))))....... ( -39.20) >sp_ag.0 2139123 120 - 2160267/2560-2680 UAGUAGCGGCGAGCGAAACGGCAGGAGGGCAAACCGAAGUGUUUACACUUCGGGGUUGUAGGACUGCGAUAUAGGAUUAAUCAUUAUAGAAGAAUUACCUGGGAAGGUAAGCCAAAGAGA .......(((..........((((....((((.((((((((....))))))))..))))....))))...........................((((((....)))))))))....... ( -36.70) >sp_mu.0 1684213 120 + 2030921/2560-2680 CAGUAGCGGCGAGCGAAGAGGCAGGAGGGCAAACCAGAGUGUUUACACUCUGGGGUUGUAGGACUGCGAUAAAGCAGCCAAGGGAAUAGAAGAAGACUCUGGGAAGAGUCGCCAGAGAGA .....(((((..((......((((....((((.((((((((....))))))))..))))....))))......)).)))...............((((((....))))))))........ ( -43.90) >consensus UAGUAGCGGCGAGCGAAACGGCAGGAGGGCAAACCGAAGUGUUUACACUUCGGGGUUGUAGGACUGCGAUAUAGAAUCAAAGAUUAUAGAAGAAUAAUCUGGGAAGAUAAGCCAAAGAGA .......(((..........((((....((((.((((((((....))))))))..))))....))))...........................((((((....)))))))))....... (-40.78 = -37.23 + -3.55) # Strand winner: reverse (1.00)

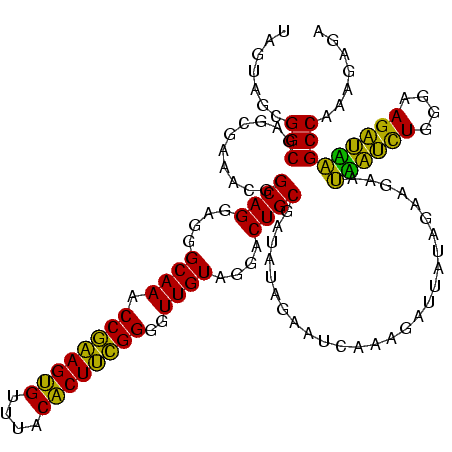
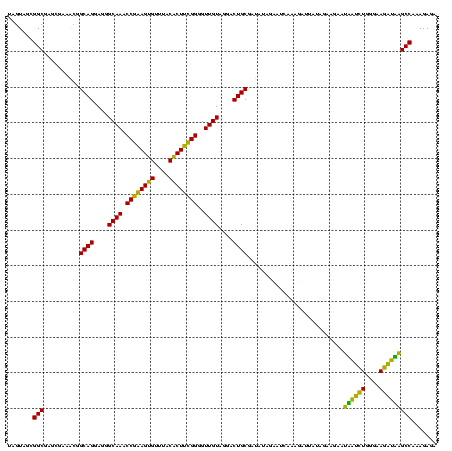
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -29.84 |
| Energy contribution | -28.29 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.980153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/2600-2720 UUGAGUCCACAUUGCAGUCCUACAACCCCGAAGAGUAAACUCUUCGGUUUGCCCUCCUGCCGUUUCGCUCGCCGCUACUAAGGCAAUCGCUUUUGCUUUCUCUUCCUGCAGCUACUUAGA ..((((....((.((((.....(((..((((((((....)))))))).))).....)))).))...))))((.((....(((((((......)))))))........)).))........ ( -30.80) >sp_ag.0 2139123 120 - 2160267/2600-2720 UUAAUCCUAUAUCGCAGUCCUACAACCCCGAAGUGUAAACACUUCGGUUUGCCCUCCUGCCGUUUCGCUCGCCGCUACUAAGGCAAUCGCUUUUGCUUUCUCUUCCUGCAGCUACUUAGA ......(((....((((.....(((..((((((((....)))))))).))).....))))......(((.((.......(((((((......)))))))........)))))....))). ( -28.66) >sp_mu.0 1684213 120 + 2030921/2600-2720 UGGCUGCUUUAUCGCAGUCCUACAACCCCAGAGUGUAAACACUCUGGUUUGCCCUCCUGCCUCUUCGCUCGCCGCUACUGAGGCAAUCGCUCUUGCUUUCUCUUCCUGCAGCUACUGAGA .((((((......))))))........((((((((....))))))))......(((.((((((...((.....))....))))))...(((...((...........)))))....))). ( -37.00) >consensus UUGAUCCUAUAUCGCAGUCCUACAACCCCGAAGUGUAAACACUUCGGUUUGCCCUCCUGCCGUUUCGCUCGCCGCUACUAAGGCAAUCGCUUUUGCUUUCUCUUCCUGCAGCUACUUAGA .............((((.....(((..((((((((....)))))))).))).....))))......(((.((.......(((((((......)))))))........)))))........ (-29.84 = -28.29 + -1.55)

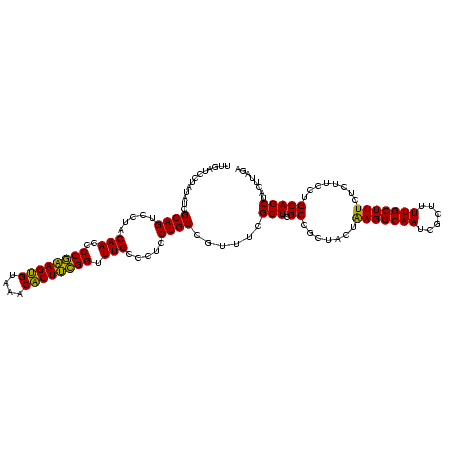
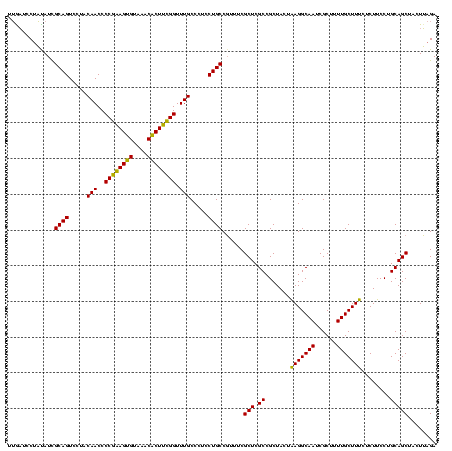
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -41.50 |
| Consensus MFE | -38.40 |
| Energy contribution | -37.07 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/2600-2720 UCUAAGUAGCUGCAGGAAGAGAAAGCAAAAGCGAUUGCCUUAGUAGCGGCGAGCGAAACGGCAGGAGGGCAAACCGAAGAGUUUACUCUUCGGGGUUGUAGGACUGCAAUGUGGACUCAA .....((.((((((((....((..((....))..)).)))..))))).))((((.(....((((....((((.((((((((....))))))))..))))....))))....).).))).. ( -41.50) >sp_ag.0 2139123 120 - 2160267/2600-2720 UCUAAGUAGCUGCAGGAAGAGAAAGCAAAAGCGAUUGCCUUAGUAGCGGCGAGCGAAACGGCAGGAGGGCAAACCGAAGUGUUUACACUUCGGGGUUGUAGGACUGCGAUAUAGGAUUAA ((((.((.((((((((....((..((....))..)).)))..))))).))..........((((....((((.((((((((....))))))))..))))....))))....))))..... ( -37.90) >sp_mu.0 1684213 120 + 2030921/2600-2720 UCUCAGUAGCUGCAGGAAGAGAAAGCAAGAGCGAUUGCCUCAGUAGCGGCGAGCGAAGAGGCAGGAGGGCAAACCAGAGUGUUUACACUCUGGGGUUGUAGGACUGCGAUAAAGCAGCCA ((((....(((............)))........(((((((....((.....))...)))))))))))((((.((((((((....))))))))..)))).((.((((......)))))). ( -45.10) >consensus UCUAAGUAGCUGCAGGAAGAGAAAGCAAAAGCGAUUGCCUUAGUAGCGGCGAGCGAAACGGCAGGAGGGCAAACCGAAGUGUUUACACUUCGGGGUUGUAGGACUGCGAUAUAGAAUCAA .....((.((((((((....((..((....))..)).)))..))))).))..........((((....((((.((((((((....))))))))..))))....))))............. (-38.40 = -37.07 + -1.33) # Strand winner: reverse (1.00)

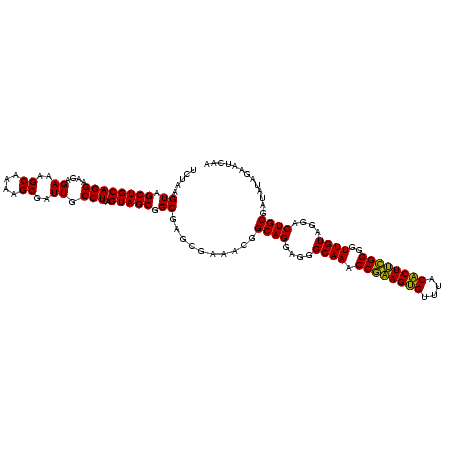
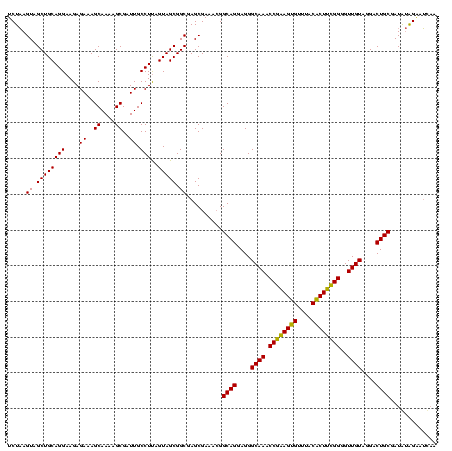
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -28.53 |
| Consensus MFE | -29.13 |
| Energy contribution | -26.47 |
| Covariance contribution | -2.66 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.22 |
| Structure conservation index | 1.02 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/2680-2800 AGGCAAUCGCUUUUGCUUUCUCUUCCUGCAGCUACUUAGAUGUUUCAGUUCACUGCGUCUUCCUCCUCACAUCCUUAACAGAUGUGGGUAACAGGUAUUACCUGUUGGGUUCCCCCAUUC ((((((......)))))).........((((..(((..((....)))))...)))).........((((((((.......)))))))).((((((.....))))))(((....))).... ( -30.80) >sp_ag.0 2139123 120 - 2160267/2680-2800 AGGCAAUCGCUUUUGCUUUCUCUUCCUGCAGCUACUUAGAUGUUUCAGUUCACUGCGUCUUCCUUCUCAUAUCCUUAACAGAUAUGGAUACUAGUCAUUAACUAGUGGGUUCCCCCAUUC ((((((......)))))).........((((..(((..((....)))))...))))........((.((((((.......)))))))).((((((.....))))))(((....))).... ( -24.20) >sp_mu.0 1684213 120 + 2030921/2680-2800 AGGCAAUCGCUCUUGCUUUCUCUUCCUGCAGCUACUGAGAUGUUUCAGUUCACUGCGUCUUCCUUCUCAUAACCUUAACAGUUAUGGAUAACAGGCAUUACCUGUUGGGUUCCCCCAUUC ((((....((....))...........((((..((((((....))))))...))))))))....((.((((((.......)))))))).((((((.....))))))(((....))).... ( -30.60) >consensus AGGCAAUCGCUUUUGCUUUCUCUUCCUGCAGCUACUUAGAUGUUUCAGUUCACUGCGUCUUCCUUCUCAUAUCCUUAACAGAUAUGGAUAACAGGCAUUACCUGUUGGGUUCCCCCAUUC ((((....((....))...........((((..(((..((....)))))...))))))))....((.((((((.......)))))))).((((((.....))))))(((....))).... (-29.13 = -26.47 + -2.66)

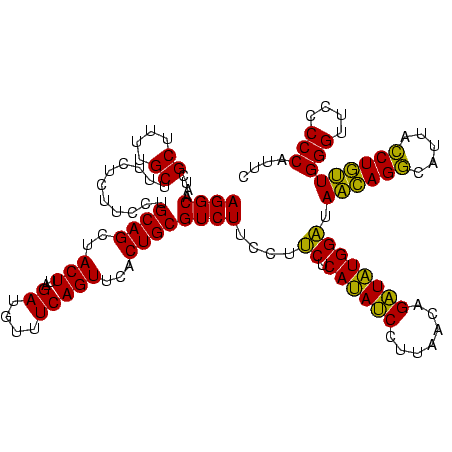
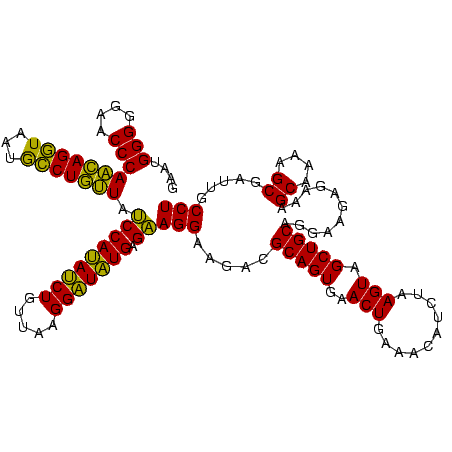
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -36.87 |
| Consensus MFE | -36.95 |
| Energy contribution | -34.07 |
| Covariance contribution | -2.88 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.37 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/2680-2800 GAAUGGGGGAACCCAACAGGUAAUACCUGUUACCCACAUCUGUUAAGGAUGUGAGGAGGAAGACGCAGUGAACUGAAACAUCUAAGUAGCUGCAGGAAGAGAAAGCAAAAGCGAUUGCCU ...((((....))))..(((((((.(((((..(((((((((.....))))))).)).(((.(...(((....)))...).)))........)))))........((....)).))))))) ( -38.90) >sp_ag.0 2139123 120 - 2160267/2680-2800 GAAUGGGGGAACCCACUAGUUAAUGACUAGUAUCCAUAUCUGUUAAGGAUAUGAGAAGGAAGACGCAGUGAACUGAAACAUCUAAGUAGCUGCAGGAAGAGAAAGCAAAAGCGAUUGCCU ....(((....)))(((((((...))))))).(((((((((.....))))))).))(((.....(((((..(((..........))).)))))...........((....)).....))) ( -31.80) >sp_mu.0 1684213 120 + 2030921/2680-2800 GAAUGGGGGAACCCAACAGGUAAUGCCUGUUAUCCAUAACUGUUAAGGUUAUGAGAAGGAAGACGCAGUGAACUGAAACAUCUCAGUAGCUGCAGGAAGAGAAAGCAAGAGCGAUUGCCU ...((((....))))..((((((((((((((.(((((((((.....)))))))...........(((((..(((((......))))).))))).......)).))).)).)).))))))) ( -39.90) >consensus GAAUGGGGGAACCCAACAGGUAAUGCCUGUUAUCCAUAUCUGUUAAGGAUAUGAGAAGGAAGACGCAGUGAACUGAAACAUCUAAGUAGCUGCAGGAAGAGAAAGCAAAAGCGAUUGCCU ....(((....)))(((((((...))))))).(((((((((.....))))))).))(((.....(((((..(((..........))).)))))...........((....)).....))) (-36.95 = -34.07 + -2.88) # Strand winner: reverse (1.00)

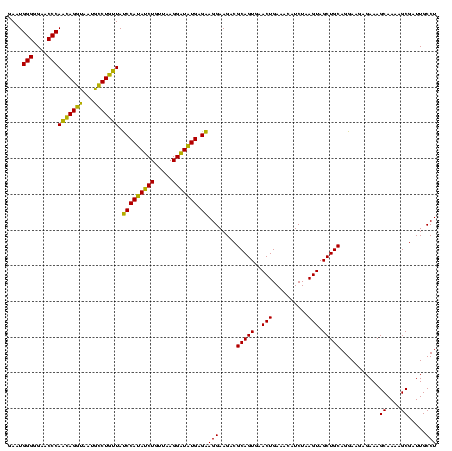
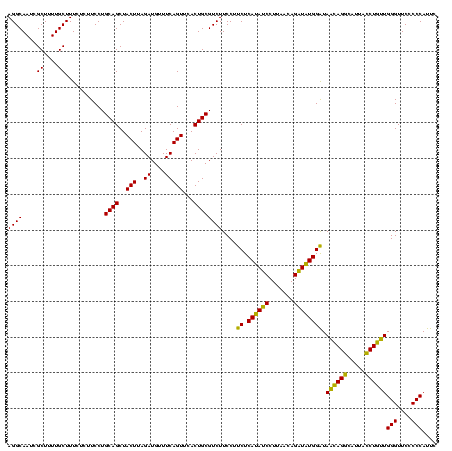
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -26.13 |
| Consensus MFE | -25.99 |
| Energy contribution | -23.67 |
| Covariance contribution | -2.32 |
| Combinations/Pair | 1.24 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645554 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/2720-2840 UGUUUCAGUUCACUGCGUCUUCCUCCUCACAUCCUUAACAGAUGUGGGUAACAGGUAUUACCUGUUGGGUUCCCCCAUUCGGAAAUCCCUGGAUCAUCGCUUACUUACAGCUACCCAAGG (((...(((.....(((...(((..((((((((.......)))))))).((((((.....))))))(((((((.......))))..))).)))....)))..))).)))........... ( -31.10) >sp_ag.0 2139123 120 - 2160267/2720-2840 UGUUUCAGUUCACUGCGUCUUCCUUCUCAUAUCCUUAACAGAUAUGGAUACUAGUCAUUAACUAGUGGGUUCCCCCAUUCGGACAUCUCUGGAUCAGCGCUUACUUACAGCUCCCCAAAG (((...(((.....((((..(((....((((((.......))))))(((((((((.....))))))(((....)))........)))...)))...))))..))).)))........... ( -23.80) >sp_mu.0 1684213 120 + 2030921/2720-2840 UGUUUCAGUUCACUGCGUCUUCCUUCUCAUAACCUUAACAGUUAUGGAUAACAGGCAUUACCUGUUGGGUUCCCCCAUUCGGAUAUCUCUGGAUCAAGGCUUACUUACAGCUCCCCAAAG .....(((....))).(((((...((.((((((.......)))))))).((((((.....))))))(((....)))(((((((....))))))).))))).................... ( -23.50) >consensus UGUUUCAGUUCACUGCGUCUUCCUUCUCAUAUCCUUAACAGAUAUGGAUAACAGGCAUUACCUGUUGGGUUCCCCCAUUCGGAAAUCUCUGGAUCAACGCUUACUUACAGCUCCCCAAAG (((...(((.....((((..(((.((.((((((.......)))))))).((((((.....))))))(((.(((.......)))...))).)))...))))..))).)))........... (-25.99 = -23.67 + -2.32)

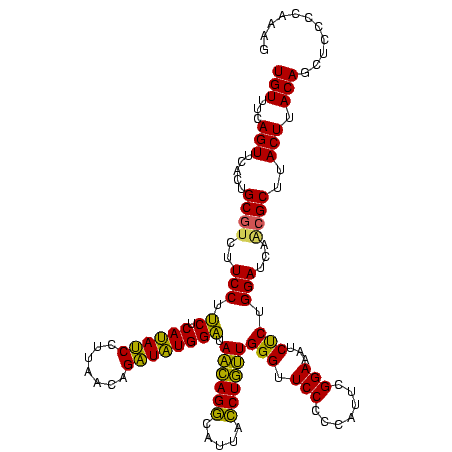
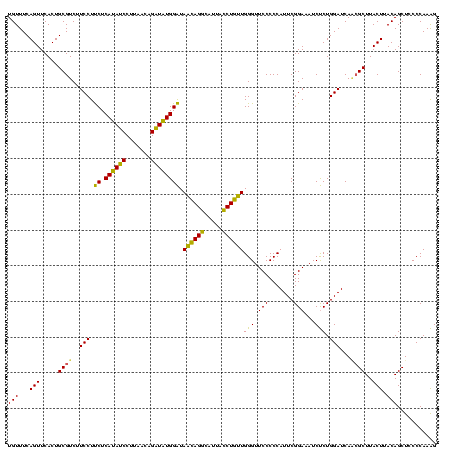
| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -41.27 |
| Consensus MFE | -42.69 |
| Energy contribution | -39.37 |
| Covariance contribution | -3.32 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.60 |
| Structure conservation index | 1.03 |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.998612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/2720-2840 CCUUGGGUAGCUGUAAGUAAGCGAUGAUCCAGGGAUUUCCGAAUGGGGGAACCCAACAGGUAAUACCUGUUACCCACAUCUGUUAAGGAUGUGAGGAGGAAGACGCAGUGAACUGAAACA ((((((((.(((.......)))....)))))))).(((((....(((....)))((((((.....)))))).(((((((((.....))))))).)).)))))...(((....)))..... ( -48.20) >sp_ag.0 2139123 120 - 2160267/2720-2840 CUUUGGGGAGCUGUAAGUAAGCGCUGAUCCAGAGAUGUCCGAAUGGGGGAACCCACUAGUUAAUGACUAGUAUCCAUAUCUGUUAAGGAUAUGAGAAGGAAGACGCAGUGAACUGAAACA (((((((.(((.((......)))))..)))))))...(((....(((....)))(((((((...))))))).(((((((((.....))))))).)).))).....(((....)))..... ( -37.70) >sp_mu.0 1684213 120 + 2030921/2720-2840 CUUUGGGGAGCUGUAAGUAAGCCUUGAUCCAGAGAUAUCCGAAUGGGGGAACCCAACAGGUAAUGCCUGUUAUCCAUAACUGUUAAGGUUAUGAGAAGGAAGACGCAGUGAACUGAAACA (((((((..(((.......))).....)))))))...(((....(((....)))((((((.....)))))).(((((((((.....))))))).)).))).....(((....)))..... ( -37.90) >consensus CUUUGGGGAGCUGUAAGUAAGCGAUGAUCCAGAGAUAUCCGAAUGGGGGAACCCAACAGGUAAUGCCUGUUAUCCAUAUCUGUUAAGGAUAUGAGAAGGAAGACGCAGUGAACUGAAACA (((((((..(((.......))).....)))))))...(((....(((....)))(((((((...))))))).(((((((((.....))))))).)).))).....(((....)))..... (-42.69 = -39.37 + -3.32) # Strand winner: reverse (1.00)

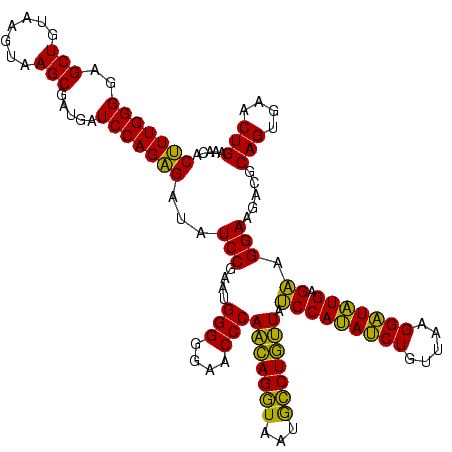
| Location | 1,790,020 – 1,790,124 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.58 |
| Mean single sequence MFE | -34.87 |
| Consensus MFE | -28.60 |
| Energy contribution | -27.93 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 104 + 2038615/3120-3240 UUUUC----------------AAUGUGCAUUACUUGGUGAUCUCUCACCAAUGGAGCCUAGCGGGAUCGAACCGCUGACCUCCUGCGUGCAAAGCAGGCGCUCUCCCAGCUGAGCUAAGG .....----------------............(((((((....))))))).((((..((((((.......))))))..)))).((.(((...))).))((((........))))..... ( -35.60) >sp_ag.0 2139123 102 - 2160267/3120-3240 UUUUC----------------AAUGAACA--AUUUGAACCUUUCGAUUCAAUGGAGCCUAGCGGGAUCGAACCGCUGACCUCCUGCGUGCAAAGCAGGCGCUCUCCCAGCUGAGCUAAGG ..(((----------------((......--..)))))((((..........((((..((((((.......))))))..)))).((.(((...))).))((((........)))).)))) ( -31.30) >sp_mu.0 1684213 120 + 2030921/3120-3240 UCUUCUACAGAAGUAUUCGCAAGCGAACCGUCUGUUAGUAUCCUGUUUUAAUGGAGCCUAGCGGGAUCGAACCGCUGACCUCCUGCGUGCAAAGCAGGCGCUCUCCCAGCUGAGCUAAGG ....(((((((....((((....))))...)))).)))...((((((((.((((((..((((((.......))))))..))))...))..)))))))).((((........))))..... ( -37.70) >consensus UUUUC________________AAUGAACA_UAUUUGAACAUCUCGAUUCAAUGGAGCCUAGCGGGAUCGAACCGCUGACCUCCUGCGUGCAAAGCAGGCGCUCUCCCAGCUGAGCUAAGG ..................................((((........))))..((((..((((((.......))))))..)))).((.(((...))).))((((........))))..... (-28.60 = -27.93 + -0.66)

| Location | 1,790,020 – 1,790,124 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.58 |
| Mean single sequence MFE | -37.30 |
| Consensus MFE | -31.06 |
| Energy contribution | -31.73 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 104 + 2038615/3120-3240 CCUUAGCUCAGCUGGGAGAGCGCCUGCUUUGCACGCAGGAGGUCAGCGGUUCGAUCCCGCUAGGCUCCAUUGGUGAGAGAUCACCAAGUAAUGCACAUU----------------GAAAA .....((..(((.(((......))))))..))..(((((((.(.(((((.......))))).).)))).(((((((....)))))))....))).....----------------..... ( -36.40) >sp_ag.0 2139123 102 - 2160267/3120-3240 CCUUAGCUCAGCUGGGAGAGCGCCUGCUUUGCACGCAGGAGGUCAGCGGUUCGAUCCCGCUAGGCUCCAUUGAAUCGAAAGGUUCAAAU--UGUUCAUU----------------GAAAA .((((((...)))))).((((((.(((...))).)).((((.(.(((((.......))))).).)))).(((((((....)))))))..--.))))...----------------..... ( -37.70) >sp_mu.0 1684213 120 + 2030921/3120-3240 CCUUAGCUCAGCUGGGAGAGCGCCUGCUUUGCACGCAGGAGGUCAGCGGUUCGAUCCCGCUAGGCUCCAUUAAAACAGGAUACUAACAGACGGUUCGCUUGCGAAUACUUCUGUAGAAGA (((..((((........))))((.(((...))).)).((((.(.(((((.......))))).).))))........)))...(((.((((..(((((....)))))...))))))).... ( -37.80) >consensus CCUUAGCUCAGCUGGGAGAGCGCCUGCUUUGCACGCAGGAGGUCAGCGGUUCGAUCCCGCUAGGCUCCAUUGAAACGAGAUAAUCAAAUA_UGUUCAUU________________GAAAA .((((((...)))))).((((((.(((...))).)).((((.(.(((((.......))))).).)))).(((((((....))))))).....))))........................ (-31.06 = -31.73 + 0.68) # Strand winner: reverse (0.89)


| Location | 1,790,020 – 1,790,140 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.44 |
| Mean single sequence MFE | -34.77 |
| Consensus MFE | -32.20 |
| Energy contribution | -31.53 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.676647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sp_pn.0 1790020 120 + 2038615/3144-3264 GUGCAUUACUUGGUGAUCUCUCACCAAUGGAGCCUAGCGGGAUCGAACCGCUGACCUCCUGCGUGCAAAGCAGGCGCUCUCCCAGCUGAGCUAAGGCCCCACAAGACCUCUCAAGACUAA (((......(((((((....))))))).((.(((((((((.......))))).....(((((.......))))).((((........))))..))))))))).................. ( -39.50) >sp_ag.0 2139123 118 - 2160267/3144-3264 GAACA--AUUUGAACCUUUCGAUUCAAUGGAGCCUAGCGGGAUCGAACCGCUGACCUCCUGCGUGCAAAGCAGGCGCUCUCCCAGCUGAGCUAAGGCCCCACAAGACCUCUCAAAACUAA .....--..(((((.(....).)))))(((.(((((((((.......))))).....(((((.......))))).((((........))))..)))).)))................... ( -32.60) >sp_mu.0 1684213 120 + 2030921/3144-3264 GAACCGUCUGUUAGUAUCCUGUUUUAAUGGAGCCUAGCGGGAUCGAACCGCUGACCUCCUGCGUGCAAAGCAGGCGCUCUCCCAGCUGAGCUAAGGCCCCACUUACCCUCUCAAAACUAA .....((((........((((((((.((((((..((((((.......))))))..))))...))..)))))))).((((........))))..))))....................... ( -32.20) >consensus GAACA_UAUUUGAACAUCUCGAUUCAAUGGAGCCUAGCGGGAUCGAACCGCUGACCUCCUGCGUGCAAAGCAGGCGCUCUCCCAGCUGAGCUAAGGCCCCACAAGACCUCUCAAAACUAA ..........((((........)))).(((.(((((((((.......))))).....(((((.......))))).((((........))))..)))).)))................... (-32.20 = -31.53 + -0.66)

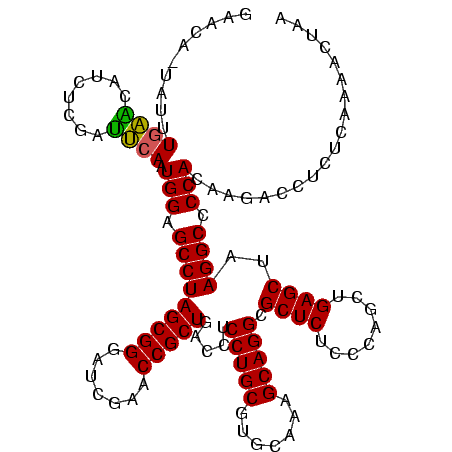
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:51:04 2006