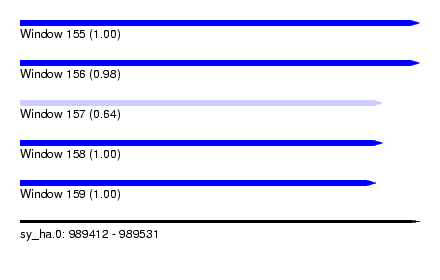
| Sequence ID | sy_ha.0 |
|---|---|
| Location | 989,412 – 989,531 |
| Length | 119 |
| Max. P | 0.999042 |

| Location | 989,412 – 989,531 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.03 |
| Mean single sequence MFE | -34.17 |
| Consensus MFE | -27.42 |
| Energy contribution | -26.87 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.14 |
| Mean z-score | -5.66 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.996252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 989412 119 + 2685015/0-120 UAAAACAUAAAAAGACGCCCCAAUUUAUGAUUGGGACGUCUAUAAGUCCCAUUCAAUGAGAAAUAGGACUUAAA-CAUUUAGUAACAGAUAAUUACUAAAGUUCCAGCCCUUGCGAGUAU ............(((((.(((((((...))))))).))))).(((((((..(((.....)))...)))))))((-(.((((((((.......)))))))))))...((....))...... ( -29.60) >sy_au.0 708422 115 - 2809422/0-120 UAAAACAUAAAAAAACGUCCCAAU--AUUAUUGGGACGUUUACAAGUCCCAUUCAAACAGAAAUAGGACUUAAAGCGUUUAGUAA---AUAUUUACUAACGUUCGAGUCCUAAAGAGUAU ............((((((((((((--...))))))))))))......................((((((((..((((((.(((((---....))))))))))).))))))))........ ( -34.80) >sy_sa.0 858419 116 + 2516575/0-120 UGAAAAUAAAAAAAGUGUCCCAAUAAAUUAUUGGGACACUUUAAAGCCCCAUUCAACUAGAAUAAGGACUUAAA-CGUUUAGUAGU--UUCUUUACUAAAGUUCCAGUCCU-AUGAGCAU ...........((((((((((((((...))))))))))))))...((...((((.....)))).((((((..((-(.((((((((.--....)))))))))))..))))))-....)).. ( -38.10) >consensus UAAAACAUAAAAAAACGUCCCAAU__AUUAUUGGGACGUUUAAAAGUCCCAUUCAAACAGAAAUAGGACUUAAA_CGUUUAGUAA___AUAUUUACUAAAGUUCCAGUCCU_AAGAGUAU ............((((((((((((.....)))))))))))).......................((((((..((...((((((((.......)))))))).))..))))))......... (-27.42 = -26.87 + -0.55)

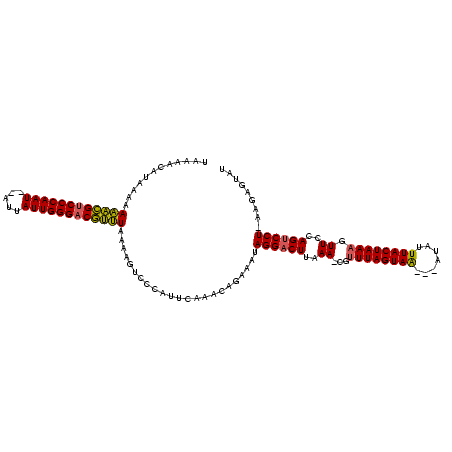

| Location | 989,412 – 989,531 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.03 |
| Mean single sequence MFE | -40.67 |
| Consensus MFE | -32.67 |
| Energy contribution | -31.57 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.15 |
| Mean z-score | -6.23 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.983929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 989412 119 + 2685015/0-120 AUACUCGCAAGGGCUGGAACUUUAGUAAUUAUCUGUUACUAAAUG-UUUAAGUCCUAUUUCUCAUUGAAUGGGACUUAUAGACGUCCCAAUCAUAAAUUGGGGCGUCUUUUUAUGUUUUA ...(((....)))...(((((((((((((.....))))))))).)-)))(((((((((((......)))))))))))..((((((((((((.....))))))))))))............ ( -44.10) >sy_au.0 708422 115 - 2809422/0-120 AUACUCUUUAGGACUCGAACGUUAGUAAAUAU---UUACUAAACGCUUUAAGUCCUAUUUCUGUUUGAAUGGGACUUGUAAACGUCCCAAUAAU--AUUGGGACGUUUUUUUAUGUUUUA ........(((((((..(((((((((((....---))))).)))).))..)))))))............((((((.((.((((((((((((...--))))))))))))...)).)))))) ( -35.70) >sy_sa.0 858419 116 + 2516575/0-120 AUGCUCAU-AGGACUGGAACUUUAGUAAAGAA--ACUACUAAACG-UUUAAGUCCUUAUUCUAGUUGAAUGGGGCUUUAAAGUGUCCCAAUAAUUUAUUGGGACACUUUUUUUAUUUUCA ..((((..-((((((.(((((((((((.....--..))))))).)-))).))))))(((((.....)))))))))...((((((((((((((...))))))))))))))........... ( -42.20) >consensus AUACUCAU_AGGACUGGAACUUUAGUAAAUAU___UUACUAAACG_UUUAAGUCCUAUUUCUAAUUGAAUGGGACUUAUAAACGUCCCAAUAAU__AUUGGGACGUUUUUUUAUGUUUUA ...(((...((((((..((.((((((((.......))))))))...))..))))))..(((.....))).)))......((((((((((((.....))))))))))))............ (-32.67 = -31.57 + -1.10) # Strand winner: reverse (1.00)

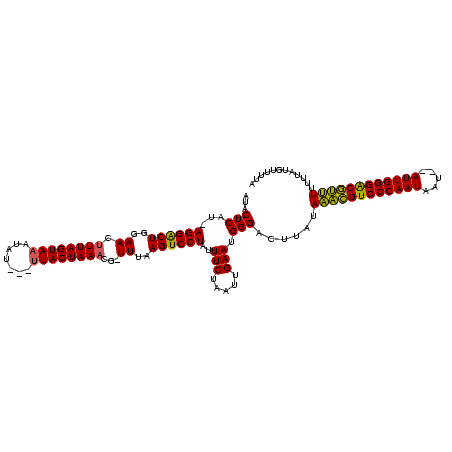
| Location | 989,412 – 989,520 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.01 |
| Mean single sequence MFE | -24.27 |
| Consensus MFE | -12.36 |
| Energy contribution | -12.70 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.51 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 989412 108 + 2685015/40-160 UAUAAGUCCCAUUCAAUGAGAAAUAGGACUUAAA-CAUUUAGUAACAGAUAAUUACUAAAGUUCCAGCCCUUGCGAGUAUAAUAAAAUAAUUA----------AAUUGUCAGUUGG-GUU ..(((((((..(((.....)))...)))))))((-(.((((((((.......)))))))))))(((((....((((...((((......))))----------..))))..)))))-... ( -21.30) >sy_au.0 708422 114 - 2809422/40-160 UACAAGUCCCAUUCAAACAGAAAUAGGACUUAAAGCGUUUAGUAA---AUAUUUACUAACGUUCGAGUCCUAAAGAGUAUAAUAAAAUAAUAAUA---UUUGAAGUUGUCGGUUGAUGUU ............((((...((..((((((((..((((((.(((((---....))))))))))).))))))))..((((((.((......)).)))---))).......))..)))).... ( -22.70) >sy_sa.0 858419 116 + 2516575/40-160 UUAAAGCCCCAUUCAACUAGAAUAAGGACUUAAA-CGUUUAGUAGU--UUCUUUACUAAAGUUCCAGUCCU-AUGAGCAUAAUAAAAUGACUAAAAUAUUUGAAGUUGUUGGUUGAGCUU ...((((((((..(((((.(((((((((((..((-(.((((((((.--....)))))))))))..))))))-...(((((......))).))....)))))..))))).)))..).)))) ( -28.80) >consensus UAAAAGUCCCAUUCAAACAGAAAUAGGACUUAAA_CGUUUAGUAA___AUAUUUACUAAAGUUCCAGUCCU_AAGAGUAUAAUAAAAUAAUUA_A___UUUGAAGUUGUCGGUUGA_GUU ............((((((......((((((..((...((((((((.......)))))))).))..))))))...............((((((...........)))))).)))))).... (-12.36 = -12.70 + 0.34)

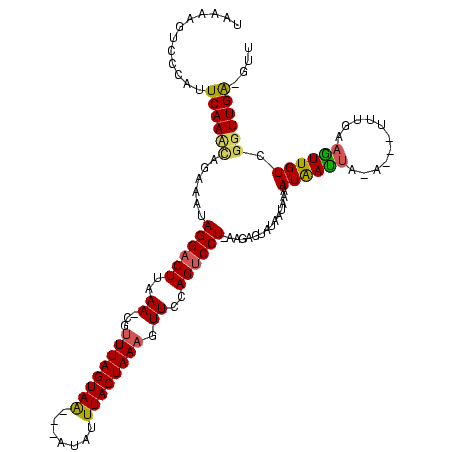
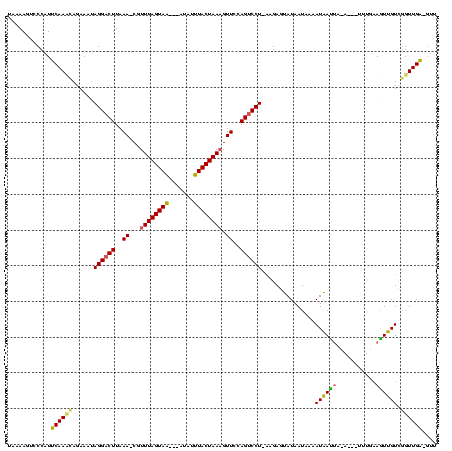
| Location | 989,412 – 989,520 |
|---|---|
| Length | 108 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.01 |
| Mean single sequence MFE | -25.93 |
| Consensus MFE | -14.06 |
| Energy contribution | -14.07 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.54 |
| SVM decision value | 3.34 |
| SVM RNA-class probability | 0.999042 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 989412 108 + 2685015/40-160 AAC-CCAACUGACAAUU----------UAAUUAUUUUAUUAUACUCGCAAGGGCUGGAACUUUAGUAAUUAUCUGUUACUAAAUG-UUUAAGUCCUAUUUCUCAUUGAAUGGGACUUAUA ...-(((..........----------((((......))))..(((....))).)))((((((((((((.....))))))))).)-))((((((((((((......)))))))))))).. ( -26.80) >sy_au.0 708422 114 - 2809422/40-160 AACAUCAACCGACAACUUCAAA---UAUUAUUAUUUUAUUAUACUCUUUAGGACUCGAACGUUAGUAAAUAU---UUACUAAACGCUUUAAGUCCUAUUUCUGUUUGAAUGGGACUUGUA .(((....(((.....((((((---((....(((......))).....(((((((..(((((((((((....---))))).)))).))..)))))))....)))))))))))....))). ( -24.20) >sy_sa.0 858419 116 + 2516575/40-160 AAGCUCAACCAACAACUUCAAAUAUUUUAGUCAUUUUAUUAUGCUCAU-AGGACUGGAACUUUAGUAAAGAA--ACUACUAAACG-UUUAAGUCCUUAUUCUAGUUGAAUGGGGCUUUAA ((((((...((((...............((.(((......)))))...-((((((.(((((((((((.....--..))))))).)-))).)))))).......))))....))))))... ( -26.80) >consensus AAC_UCAACCGACAACUUCAAA___U_UAAUUAUUUUAUUAUACUCAU_AGGACUGGAACUUUAGUAAAUAU___UUACUAAACG_UUUAAGUCCUAUUUCUAAUUGAAUGGGACUUAUA ........(((......................................((((((..((.((((((((.......))))))))...))..))))))..(((.....))))))........ (-14.06 = -14.07 + 0.01) # Strand winner: reverse (0.81)

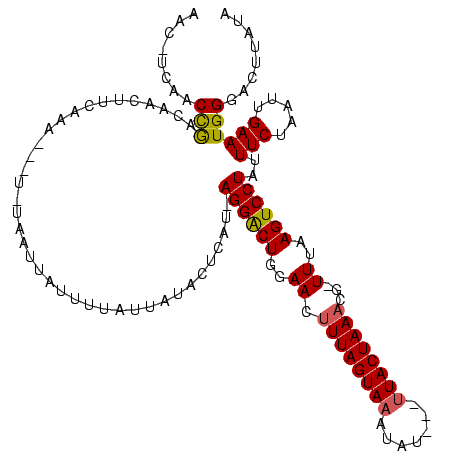
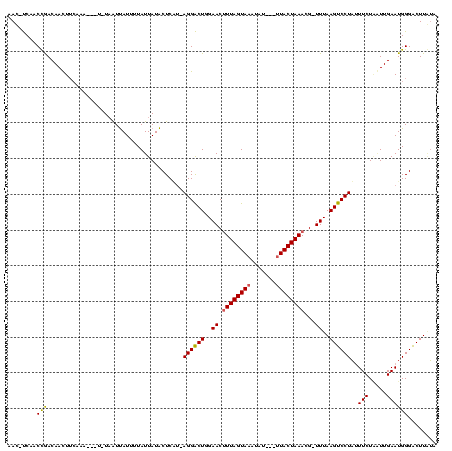
| Location | 989,412 – 989,518 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 68.27 |
| Mean single sequence MFE | -18.90 |
| Consensus MFE | -14.48 |
| Energy contribution | -14.93 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.77 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>sy_ha.0 989412 106 + 2685015/80-200 AA--GGAAGGAGUAUAUUAAAAUGACAAAUCAA-ACAUAUAAC-CCAACUGACAAUU----------UAAUUAUUUUAUUAUACUCGCAAGGGCUGGAACUUUAGUAAUUAUCUGUUACU ..--.(((((((((((.((((((((........-.........-.............----------...)))))))).)))))))((....)).....))))((((((.....)))))) ( -16.21) >sy_au.0 708422 112 - 2809422/80-200 AC--GGAAGGAGUAUAAUAAAAUGCUUAAUCAAUAUACUGAACAUCAACCGACAACUUCAAA---UAUUAUUAUUUUAUUAUACUCUUUAGGACUCGAACGUUAGUAAAUAU---UUACU .(--..((((((((((((((((((.....(((......))).........((.....))...---......))))))))))))))))))..)...........(((((....---))))) ( -18.70) >sy_sa.0 858419 117 + 2516575/80-200 UAUCGAGAGGAGUAUAAUAAAAUGACCAAACAUACAACGCAAGCUCAACCAACAACUUCAAAUAUUUUAGUCAUUUUAUUAUGCUCAU-AGGACUGGAACUUUAGUAAAGAA--ACUACU ..((.((..(((((((((((((((((...........(....).................((....)).)))))))))))))))))..-....)).))....((((......--)))).. ( -21.80) >consensus AA__GGAAGGAGUAUAAUAAAAUGACAAAUCAA_AAACACAAC_UCAACCGACAACUUCAAA___U_UAAUUAUUUUAUUAUACUCAU_AGGACUGGAACUUUAGUAAAUAU___UUACU .........(((((((((((((((((...........................................)))))))))))))))))......(((((....))))).............. (-14.48 = -14.93 + 0.45) # Strand winner: reverse (0.97)

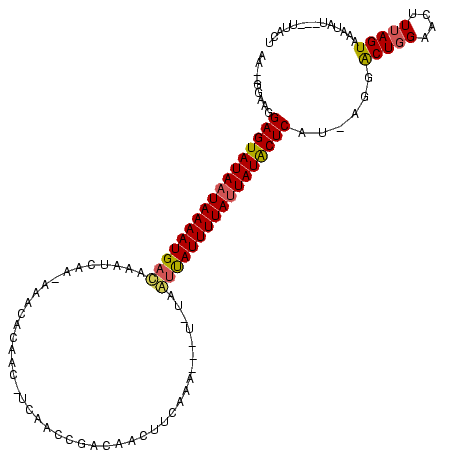
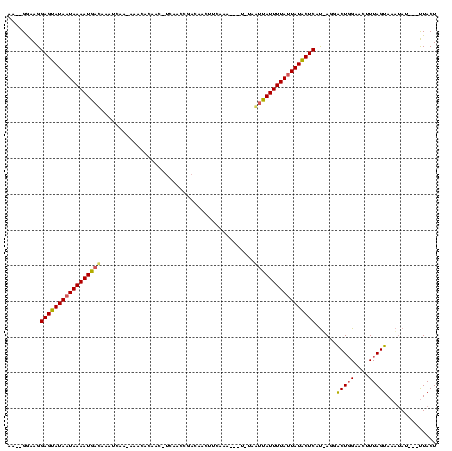
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 18:52:47 2006